Overview
The 16S rRNA gene sequence is frequently used to identify and profile membership of bacterial communities. The utility of studying 16S for microbial classification and phylogenetic analysis was demonstrated over 40 years ago by Carl Woese and its impact has only increased over time. Defining and understanding microbial communities has a broad range of important applications, including human health, environmental surveys, animal welfare, and agricultural improvements.
Our myBaits 16S-Hyb Panel enriches the entire gene, rather than just one or two hypervariable regions, leading to better taxonomic resolution and less bias than 16S amplicon sequencing. The panel can also efficiently identify rare taxa that may be contributing to novel phenotypes, in conjunction with metagenomic studies. Achieve improved characterization of microbial populations in metagenomic samples more cost-effectively than shotgun sequencing alone and more accurately than 16S amplicon sequencing by combining 16S-Hyb rRNA gene enrichment with low-coverage shotgun sequencing.
- Convenient – Use the same NGS library preparation or protocol for shotgun sequencing.
- Cost-effective – Identify rare taxa in metagenomic samples with less sequence depth.
- Less bias – Avoids issues associated with amplicon-based approaches.
- Excellent resolution – All constant and hypervariable regions are represented.
- Open platform – Compatible with any NGS library platform, including long-read sequencing.
- Complete solution – Kits include all necessary reagents to perform enrichment of NGS libraries.
Want data even faster? Leverage our expert myReads® NGS services.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Performance
The myBaits 16S panel was compared to 16S amplicon and shotgun metagenomic sequencing for samples obtained from a mock community, mouse feces, and rat colon. The unenriched metagenomic sequences contained ~0.1-0.2% 16S reads; enrichment led to a 350-fold enrichment for the mock communities and 450-fold enrichment for the mouse feces and rat colon, with 60-70% of reads on target. Overall, the profile of the enriched samples using either MiSeq or HiSeq sequencing much more closely resembled the mock and shotgun profiles versus the amplicon data in all experiments.
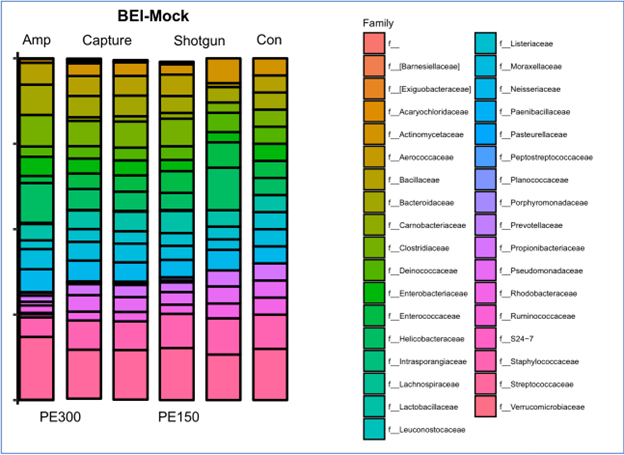
BEI Resources mock community analysis
The BEI Resources mock community contains a bacterial population of known content (Column 5) and was analyzed by 16S amplicon sequencing using MiSeq PE 300 (Column 1), myBaits 16S enrichment and sequencing using MiSeq PE 300 (Column 2) or HiSeq PE 150 (Column 3), and shotgun sequencing using HiSeq PE 150 (Column 4). Enriched and unenriched reads were trimmed and filtered, mapped against the GreenGenes database using bbmap, and analyzed. Amplicon reads were assessed using the Dada2 pipeline and GreenGenes database.
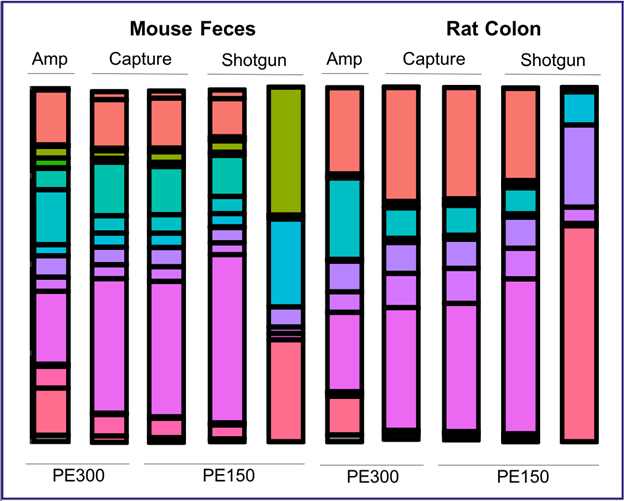
16S amplicon vs 16S-hyb vs whole shotgun sequencing
DNA extracted from mouse feces and rat colon was analyzed by 16S amplicon sequencing using MiSeq PE 300 (Columns 1,5), myBaits 16S enrichment and sequencing using MiSeq PE 300 (Columns 2,6) or HiSeq PE 150 (Columns 3,7), and shotgun sequencing using HiSeq PE 150 (Columns 4,8). Enriched and unenriched reads were trimmed and filtered, mapped against the GreenGenes database using bbmap, and analyzed. Amplicon reads were assessed using the Dada2 pipeline and GreenGenes database.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Resources
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.