Overview
This myBaits® probe set was designed by field experts and described in Systematic Biology to target 353 putative single-copy loci. Each bait was selected using a k-medoid clustering approach starting from aligned genomic sequences among highly diverse flowering plants, which makes the kit both streamlined and versatile. It boasts a large and growing bibliography, the data from which is aggregated in the phenomenal and publicly available Kew Tree of Life Explorer.
- Targets 353 single-copy loci in flowering plants.
- Compatible with fresh, herbarium, and museum samples.
- Standardized data analysis using HybPiper
- Publicly accessible database of Angiosperms353 data at Kew Tree of Life Explorer.
- Single, all-inclusive kit for target enrichment–just add your libraries.
Want data even faster? Leverage our expert myReads NGS services. We have processed thousands of samples using the Angiosperms353 probe set and are ready to help with your project.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Performance
The myBaits Expert Angiosperms-353 kit can retrieve orthologous loci across a huge taxonomic range within flowering plants, illuminating both shallow and deep evolutionary relationships.
A Universal Phylogenomics Solution for Flowering Plants
The myBaits Expert Angiosperms353 kit has been demonstrated to work across a very wide taxonomic and sample range, with excellent exonic and intronic recovery that varies depending on the species and the quality of DNA input. Some example data are shown below, but for the latest and most comprehensive results, check out the most recent Publications that have been published utilizing this kit for an even greater variety of species and sample types.
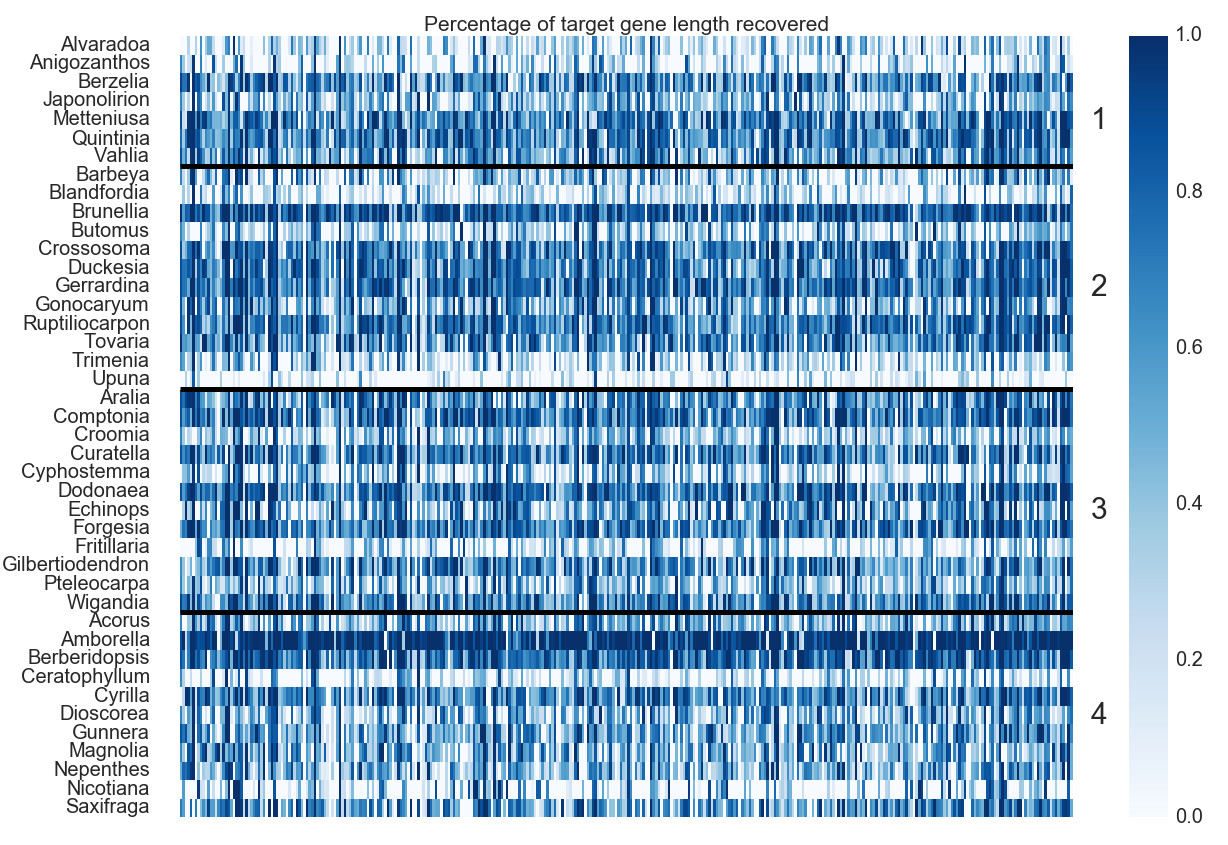
Heatmap of Gene Recovery Efficiency
“Each row is one sample, and each column is one gene. Colors indicate the percentage of the target length (calculated by the mean length of all k-medoid transcripts for each gene) recovered.” (pg 19, Johnson, Pokorny, Dodsworth et al 2019, Syst Bio). Image is Figure 3 from Johnson, Pokorny, Dodsworth et al 2019, Syst Biol.
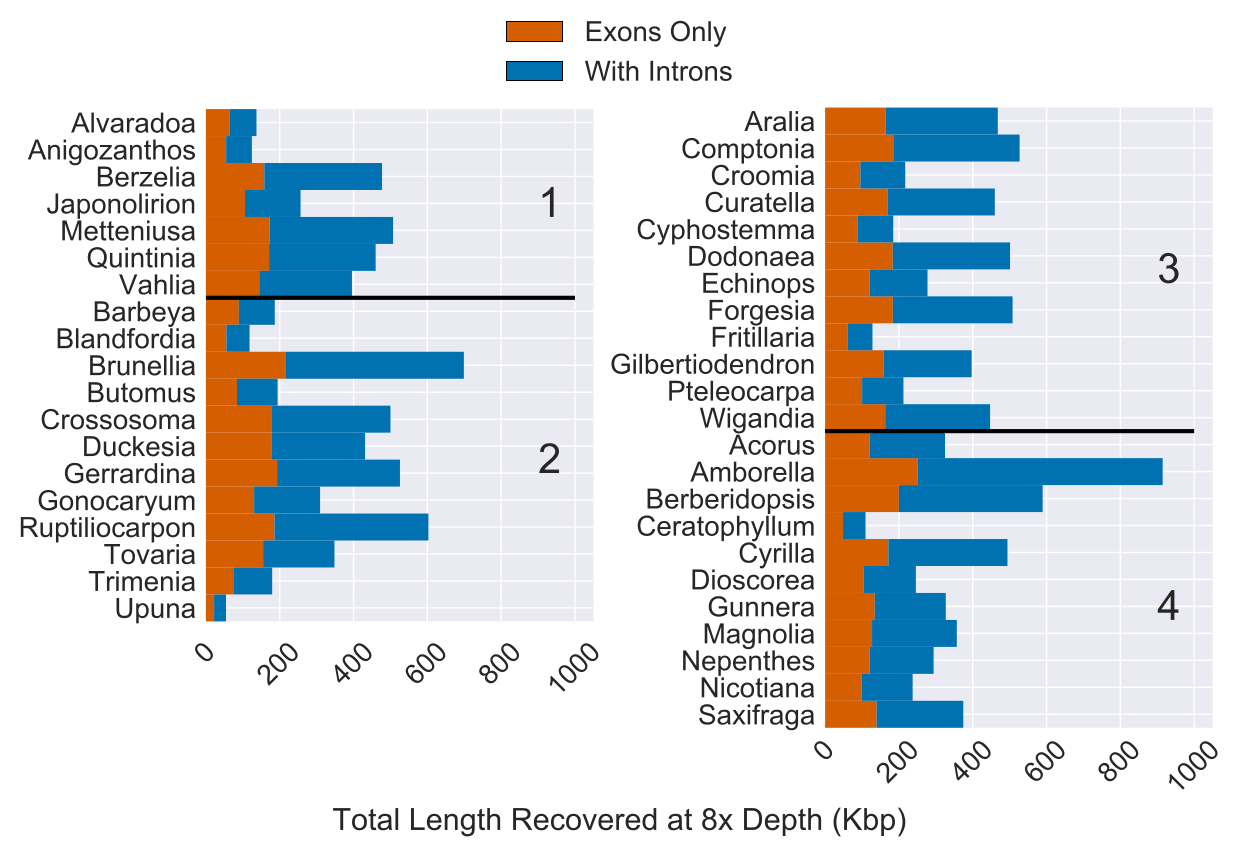
Total Length of Sequence Recovery for Both Coding and Non-coding Regions Across 353 Loci for 42 Angiosperm Species
“Reads were mapped back to either coding sequence (yellow) or coding sequence plus flanking non-coding (i.e. intron) sequence (purple)…The total length of coding sequence targeted was 260,802 bp. The median recovery of coding sequence was 137,046 bp and the median amount of non-coding sequence recovered was 216,816 bp (with at least 8x depth of coverage).” (pg 21, Johnson, Pokorny, Dodsworth et al 2019, Syst Bio). Image is modified version of Figure 4 from Johnson, Pokorny, Dodsworth et al 2019, Syst Bio.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Resources
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Publications
For more studies that utilize the Angiosperms-353 panel, search our comprehensive Publications database.
American Journal of Botany | July 1, 2021
Applications in Plant Sciences | January 1, 2021
Applications in Plant Sciences | January 1, 2021
HybPhaser: A workflow for the detection and phasing of hybrids in target capture data sets
Applications in Plant Sciences | January 1, 2021
New targets acquired: Improving locus recovery from the Angiosperms353 probe set
Applications in Plant Sciences | January 1, 2021
On the potential of Angiosperms353 for population genomic studies
Applications in Plant Sciences | April 1, 2020
Trends in Plant Science | August 30, 2019
Systematic Biology | July 1, 2019
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.