Overview
Get better data— faster— with the new Library Prep Kit for myBaits targeted sequencing.
- Premium formulas – Advanced enzyme mixes make the most of even low-input or degraded DNA samples.
- Convenient kits – Unique sizes provide maximum compatibility with myBaits capture.
- Full workflow – A complete targeted sequencing solution when paired with your myBaits kit and preferred barcoding system.
- Broad compatibility – Prep 1-500 ng high molecular weight or degraded dsDNA.
Libraries built with this kit can be paired with any appropriate myBaits Custom or Predesigned kit. Contact us today to get started on your targeted sequencing project.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Applications
- DNA samples for targeted sequencing
- Human, plant, animal, fungus, virus, or bacteria samples
- FFPE or cfDNA samples
- Degraded, museum, herbarium, or historical samples
- Hybridization capture or target enrichment
- Targeted gene or exon sequencing
- Viral, bacterial, or microbial genome sequencing
- Metagenomic or microbiome targeted sequencing
- 16S or AMR capture sequencing
- Whole exome sequencing (WES)
- Phylogenetics or evolutionary biology
- Genotyping and population genetics
Questions about whether the Library Prep Kit for myBaits is the right choice for your project? Read our FAQs, review the protocol, or reach out to talk to our experts.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Performance
Complete workflow with myBaits hybridization capture
The Library Prep Kit provides reagents to prepare NGS libraries (with user-supplied adapters) as well as for the post-capture amp step of the myBaits protocol. Available in two sizes that enable pooling multiple libraries in each myBaits reaction, the Library Prep Kit is your seamless solution to generating high-quality targeted DNA sequencing data.
Tunable to achieve your desired library size
myBaits targeted sequencing projects typically utilize insert lengths of ~300-500 bp, but many applications benefit from longer or shorter libraries. With the Library Prep Kit, you control library size while avoiding time-consuming, costly sonication-based shearing.
Handles a wide range of input DNA qualities
Generating a comprehensive, representative NGS library is the critical first step for any successful hybridization capture project. With the Library Prep Kit for myBaits, you can use the same kit even for low input, sensitive, and/or degraded DNA samples.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Resources
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
FAQs
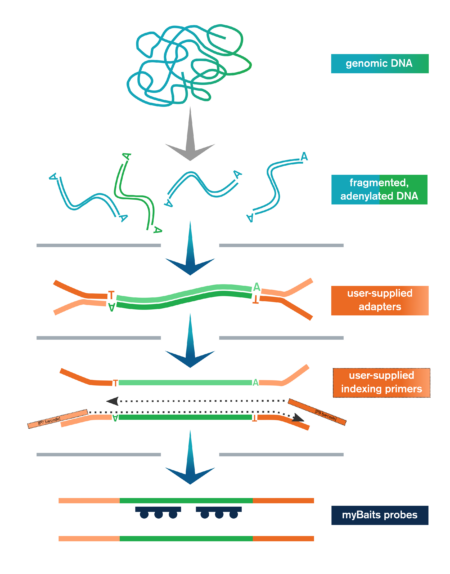
The Library Prep Kit streamlines NGS library preparation for hybridization capture with myBaits. It includes:
- Genomic or other dsDNA is enzymatically fragmented, end-polished, and adenylated
- User-supplied adapters are ligated to the end-repaired fragments
- Ligation products are purified with SPRI beads and then amplified and SPRI'd again
- Libraries are optionally pooled and taken to myBaits capture
In addition to the upstream library preparation steps, the Library Prep Kit for myBaits includes the reagents necessary for performing the post-capture amplification step of the myBaits protocol.
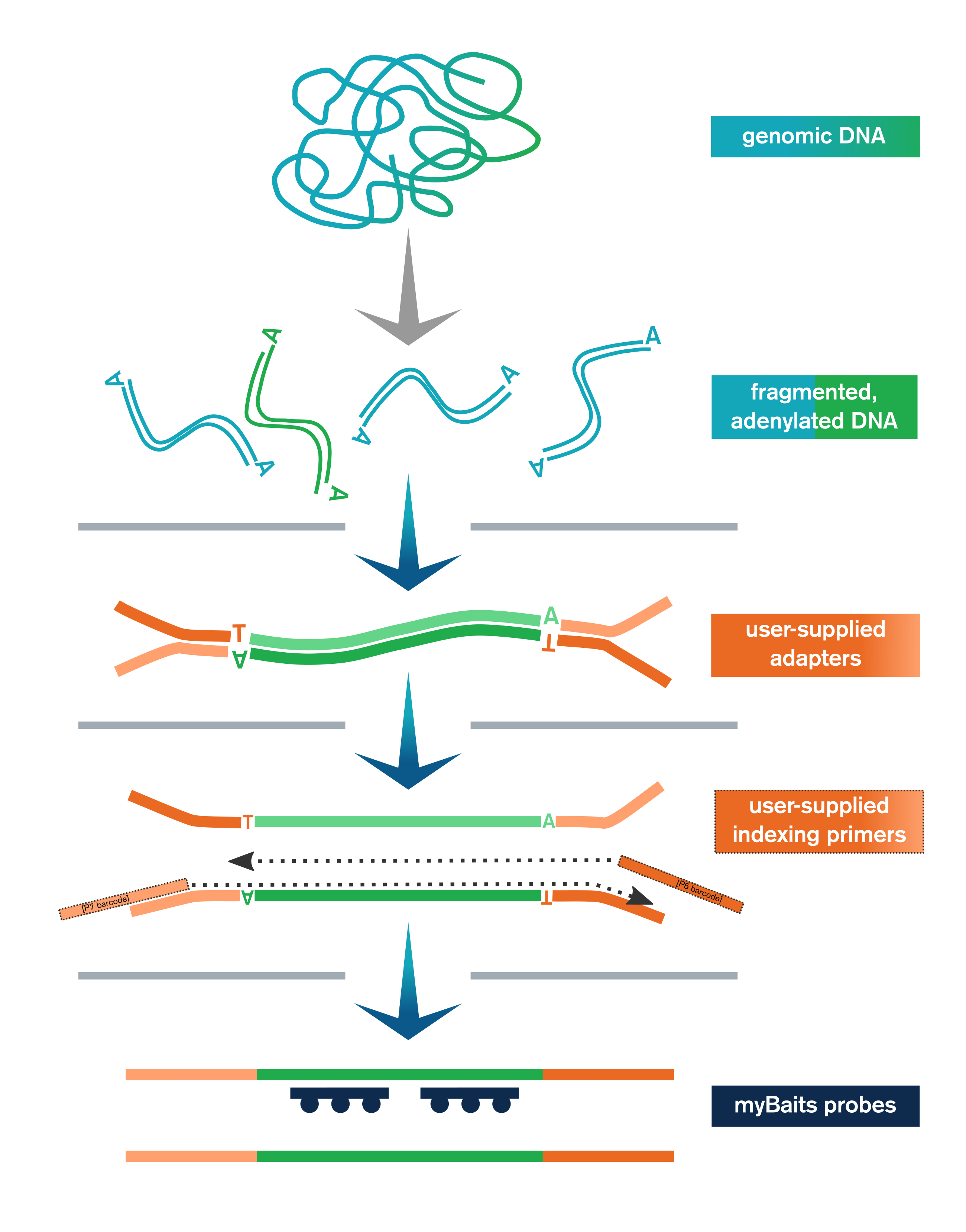
The Library Prep Kit includes reagents for key steps in NGS library preparation, including end-polishing/A-tailing, enzymatic fragmentation, ligation of user-supplied adapters, purification, and amplification (with user-supplied indexing primers, or kit-supplied universal P5/P7 primers if using full-length adapters).
In addition, the kit also provides sufficient reagents for the amplification and purification steps at the end of the myBaits capture protocol (myBaits kit sold separately).
Full details of what is included in the kit and what reagents/equipment are required to complete the protocol can be found in the kit manual.
The Library Preparation Kit for myBaits is intended to be used for the preparation of NGS libraries from DNA samples prior to hybridization capture with myBaits.
Compatible DNA samples should be or have:
- 1-500 ng genomic or other dsDNA
- <= 0.1mM EDTA storage buffer
- No viscosity or coloration
- Any length distribution; see technical recommendations for selecting frag time
Libraries made with this protocol are directly compatible with downstream myBaits enrichment (or another hybridization capture system), or alternatively can be sequenced directly in order to generate non-enriched read data.
No. myBaits hybridization capture kits have always been-- and will continue to be-- compatible with any user-provided upstream NGS library preparation system that is appropriate for a given project. This ensures flexibility in your targeted next-generation sequencing workflow.
However, for your convenience, we are now offering our own powerful kit for preparing NGS libraries from most types of DNA samples, which is relevant for many myBaits hybridization capture projects. This new product (Library Preparation Kit for myBaits) is intended to be used for the preparation of NGS libraries from dsDNA samples prior to hybridization capture with myBaits.
In addition to the upstream library preparation steps, the Library Prep Kit for myBaits includes the reagents necessary for performing the post-capture amplification step of the myBaits protocol.
The Library Prep Kit for myBaits is compatible with a wide range of pre-built or DIY adapters or primers. The adapters used must be T-overhang-containing double-stranded adapters. These can be either short (“stubby”) adapters OR full-length adapters that contain sample-specific barcodes. If using stubby adapters, indexing primers that add universal P5 and P7 priming sites are also required. See the kit manual for further technical information about adapters and primers.
We recommend using unique dual indexes. When selecting indexes, ensure that all libraries that you ever plan to co-enrich or co-sequence have unique index combos.
For your convenience, below are listed some commercially-available adapter and barcoding solutions designed for Illumina(R) short-read sequencing, but other vendors also supply compatible options.
Stubby adapters + indexing primers
- IDT
- xGen™ Stubby Adapter in 16 rxn (10005974) or 96 rxn (10005924)
- xGen™ UDI Primer Pairs in 8nt 16 rxn (10005975), 8nt Plate 1 (10005922), 10nt Plates 1-4 (10008052), 10nt Plates 1-8 (10008053), or 10nt Plates 1-16 (10008054)
- NEB
- NEBNext® Multiplex Oligos for Illumina® | 96 Unique Dual Index Primer Pairs - Set 1 (E6440S), Set 2 (E6442S), Set 3 (E6444S), Set 4 (E6446S), Set 5 (E6448S). (Note: Do not use the included "NEBNext Adaptor" unless you open the hairpin loop with USER treatment.)
Full-length adapters
- IDT
- xGen™ UDI-UMI Adapters in 16 rxn (10006914) or 96 rxn (10005903)
- NEB
- NEBNext® Multiplex Oligos for Illumina® | Unique Dual Index UMI Adaptors DNA - Set 1 (E7395S), Set 2 (E7874S), Set 3 (E7876S), Set 4 (E7878S)
The Library Prep Kit for myBaits is designed for input amounts from 1 to 500 ng. Please see the kit manual for technical recommendations.
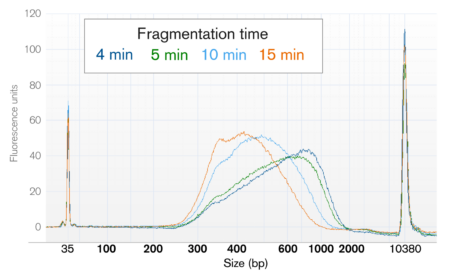
Please see the kit manual for detailed technical recommendations on the enzymatic fragmentation step. In brief, the final length of your DNA after fragmentation will depend on the (1) the length of your starting genomic DNA sample and (2) your chosen fragmentation time.
myBaits hybridization capture can tolerate NGS library insert lengths from 50bp-10Kbp. However for most myBaits applications, users should aim to generate NGS libraries with ~300-500bp length inserts (~450-650bp length libraries).
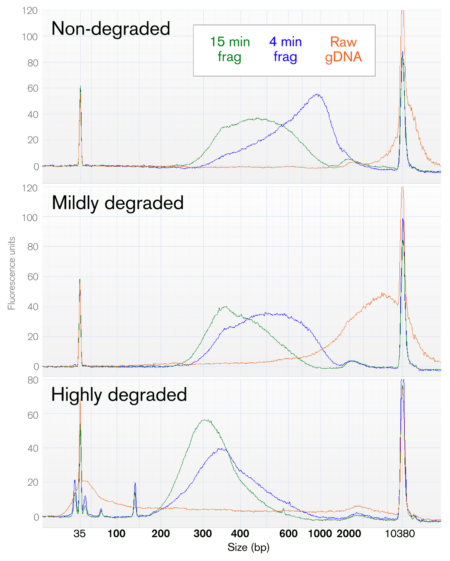
Yes, the Library Preparation Kit for myBaits is compatible with a wide range of input DNA qualities, from high-molecular weight to highly degraded.
However, some degraded sample types (such as formalin-fixed, ancient DNA, and others) may have chemical modifications, crosslinking, sequence breaks, or other types of damage that can restrict the conversion those DNA molecules into a functional NGS library. In order to be compatible with this Library Prep Kit, the target DNA molecules must be double-stranded and able to be enzymatically end-polished and adenylated (A/T overhang) to enable ligation of the sequencing adapters.
Regardless, the enzymatic fragmentation outcome will be affected by the starting length of your DNA samples. Please see the kit manual for technical recommendations for selecting the correct fragmentation approach for your project goals. In brief, the final length of DNA after fragmentation will depend on the (1) length of starting genomic DNA and (2) the chosen fragmentation time.
Yes! From our team of experts, we offer comprehensive NGS laboratory and bioinformatic services appropriate for a wide range of sample types. We offer flexible services including DNA/RNA extraction, NGS library preparation, myBaits capture, short- or long-read sequencing, and/or bioinformatic analysis.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.



 Bluesky
Bluesky