Overview
myTags Custom probes from Daicel Arbor Biosciences have been used successfully in (F)ISH experiments with a wide range of organisms such as animals, plants, and microbes – and for a variety of research applications such as 3D DNA-FISH, RNA-FISH, chromosome painting and barcoding, and much more.
Our myTags probes can be easily integrated into your existing workflows to enable (F)ISH research on chromosomal material or whole cells. With immortal and labeled options and a wide range of available synthesis scales, myTags probes are the perfect choice for any in situ hybridization experiment.
Affordable and scalable—Our proprietary oligo synthesis platform and flexible labeling options allow production of high-quality probe pools at extremely competitive pricing for any project size.
Flexible design—We can design your probes from any genome reference sequence(s), and can accommodate any cytogenetics use case, such as chromosome painting or barcoding, RNA-FISH, and much more.
Broad range of probeset sizes—Our optimized design and synthesis technologies can accommodate any size target, from a single locus to a full chromosome.
Access to expertise—Our team of expert scientists provide complimentary probe and project design assistance.
Easy to use—With ready-to-use labeled options and multiple optimized protocols available, myTags probes are ideal for new or expert cytogenetics ISH users.
Convenient options—Indexed and Standard Synthesis formats and multiple Labeling Services support any custom project cadence, from one-time to routine.
myTags probes are for research use only and are not validated for diagnostic or therapeutic purposes.
Performance
myTags Custom probes have been demonstrated to achieve highly specific targeted F(ISH) across a wide range of applications and species. Our scientists are continually innovating novel improvements to our design algorithms, labeling and use protocols, and application development to maximize performance across a variety of applications. We are always available to provide experimental design advice to maximize your F(ISH) experimental success.
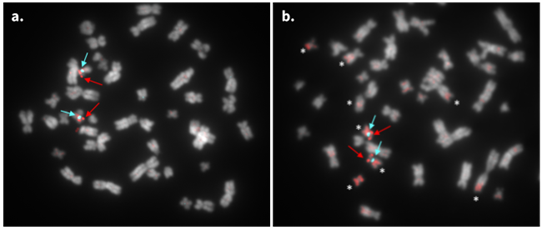
myTags probes demonstrate higher specificity compared to genomic-derived probes
Comparing myTags synthetic probes (a) versus BAC-derived probes (b) for the human TP53 locus [ATTO 550] demonstrates that the myTags probes only hybridize to the intended locus, while the BAC-derived probes also display multiple off-target hybridizations, denoted by asterisks in (b). Both panels include myTags human chromosome 17 centromere probes [ATTO 647N] and chromosomes [stained with Hoescht 33342] pseudo-colored gray.
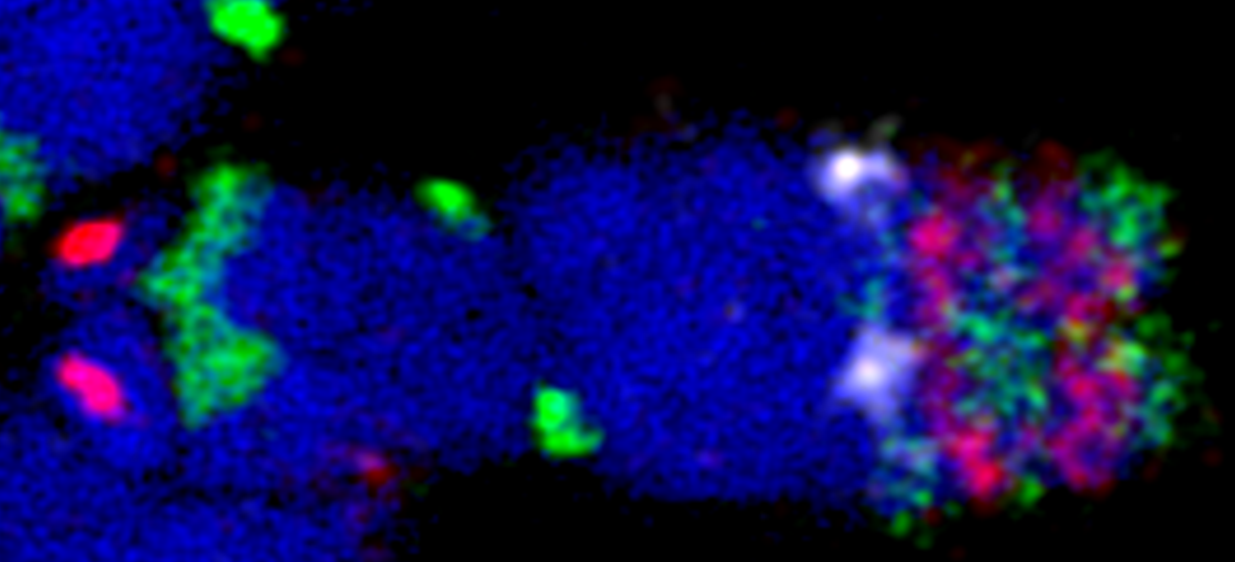
myTags probes enable super-resolution mapping of chromosomal spatial structures
(Image credit: V. Schubert.) Oligo-FISH combined with spatial super-resolution structured illumination microscopy (3D-SIM) is a useful approach for resolving helical versus non-helical arrangement of chromatin fibers. Randall et al. (2022) used this approach to confirm that the barley metaphase chromatids are formed by a helically wound ~400 nm chromatin fiber, termed chromonema.
Additionally, by measuring the volume of oligo-FISH painted regions, they were able to calculate levels of chromatin compaction. For more information, visit the original publication: Randall et al. (2022). Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes. Nucleus 13(1): 279-301
For more examples of myTags Custom probes in action, visit our myTags Featured Applications Gallery and search our Publications page.
myTags probes are for research use only and are not validated for diagnostic or therapeutic purposes.
Applications
myTags (F)ISH probes have been used successfully across a diverse range of cytogenetic applications including 3D DNA-FISH, Cryo-FISH, RNA-FISH, Oligopaints, and chromosome painting for a variety of cell types.
- Identify chromosomes with multicolor FISH or barcoded signals
- Assemble scaffolds into chromosomes with visual confirmation of their position
- Track changes in chromosome structure–translocations, duplications, deletions, etc.
- Gene mapping and gene expression
- Flow cytometry
- Fluorescent reporters
- And more
Visit our myTags Gallery page to view examples of many of these applications.
Resources
Application Note
Publication Note
Illuminate meiotic crossover events with myTags fluorescent probes for haplotyping analysis
myTags probes are for research use only and are not validated for diagnostic or therapeutic purposes.
FAQs
We may be able to accommodate other labeling options, please contact us for availability.
The number of assays per library depends on a number of factors including the probe density of your library, the size of your target region, the number of probes in the library, and the FISH protocol. Generally we recommend starting with 10pmol of labeled probes per standard FISH slide and then modifying the input amount based on the initial results.
The latest recommended protocols for both labeled and immortal probe libraries can be found in the Resources tab, but in general, myTags FISH libraries are compatible with most FISH protocols. Please contact us for specific recommendations.
We can often accommodate customer-designed probes into the myTags labeling framework. Please contact us for recommendations on the design parameters and other information before designing your probe sequences.
Yes, we can synthesize immortal probe libraries that can be labeled using the Oligopaints labeling method. Please note these probe libraries are not compatible with the standard myTags labeling protocol due to sequence requirements of the Oligopaints method.
We generally ask for up to 3-4 weeks after an order is placed to ship myTags libraries.
We recommend using the myTags labeling protocol with myTags immortal libraries for perfroming the labeling process in your own lab. If you would like to use a different protocol, one of our scientists would be happy to provide assistance to ensure success.
We offer complementary myTags Custom (F)ISH probe bioinformatic design service using our proprietary design algorithms for most types of (F)ISH projects. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will apply our advanced proprietary probe design algorithms to craft a custom probe design for your targeting needs, for any organism or application. Our scientific team has decades of experience designing custom probes for a wide variety of applications, including multi- or single locus/gene localization, chromosome painting, chromosomal indexing/barcoding, haplotyping, and more.
Only the highest specificity regions with minimal background noise and cross-reactivity for premium performance in any downstream experimental application would be selected for the probe design.
And with our transparent and straightforward scientific communication process, you always have full control and ownership over the oligo sequences for each and every myTags Custom probeset.
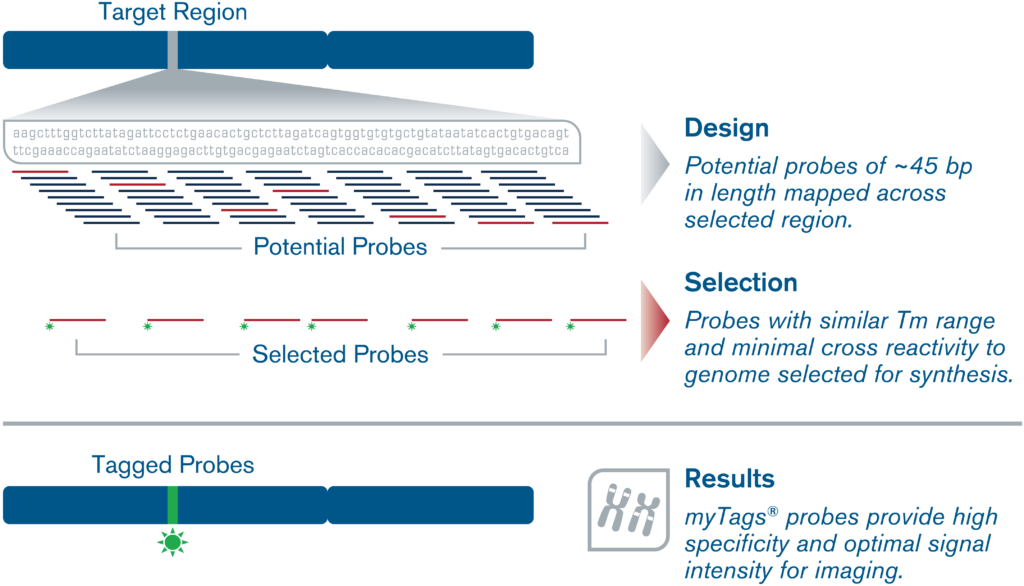
Generally we recommend probe densities between 3-10 probes per kilobase for target regions larger than 50kb. For target regions between 10-50kb, probe densities should be on the higher end of that range, and we may recommend using multiple fluorophores per probe to boost the signal.
We can work with any sequence to design probes. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will advise you on the most appropriate synthesis configuration for your desired project needs, depending on the number of individual probe sequences required for your design, and the number of individual probesets required for your experiment.
For standalone orders and/or complex probe designs requiring up to 100K+ oligos, our Single Synthesis option provides maximum value. For smaller and/or multiple designs, our new Indexed Synthesis option maximizes both flexibility and cost-effectiveness.
Regardless of the synthesis approach used, all unique probesets are delivered individually and include individual final probeset pool composition verification via next-generation sequencing.
You can choose to receive your individual probe pool(s) either as immortal amplifiable substrate(s) ready for labeling, or pair with our flexible Labeling Service to receive ready-to-use probes for your experiments.
For standalone orders and/or complex probe designs requiring up to 100K+ oligos, our Single Synthesis option provides maximum value. For smaller and/or multiple designs, our new Indexed Synthesis option maximizes both flexibility and cost-effectiveness. All probesets are delivered individually (200 ng minimum yield) and include composition verification via next-generation sequencing to confirm the quality of probe synthesis.
Arbor’s experts will assist you with determining which of our product configurations is most appropriate for your experimental goals and target type(s). You will have full transparency and control over the entire process. Contact us today to get started with your next project and experience the many benefits of using synthetic custom (F)ISH probes.
With Arbor’s flexible oligo synthesis manufacturing technology, we offer two configurations:
Single Synthesis
- Oligo pools containing 1 probeset design
- Can contain up to 108K unique probe sequences per design (larger pools may be available)
- 6 different sizes, depending on total number of probes in design: 1-1.8K, 1.8K-4K, 4K-27K, 27K-54K, 54K-81K, or 81K-108K probe sequences
Indexed Synthesis
- Oligo pools containing 2-10 probeset designs per synthesis event (up to 27K total oligos)
- If >10 designs and/or >27K oligos are needed, add additional oligo pools to scale up
- Oligos are separated into individual probesets via PCR deconvolution
Note: With Indexed Synthesis, short index sequences remain present on the final probe oligos, but are short and not expected to interfere with downstream (F)ISH protocols.
For all myTags Custom orders, final oligo pool composition is verified by next-generation sequencing (NGS), and oligos are delivered as individual probesets.
Add on myTags Labeling Services to get your (F)ISH experiments started quickly!
myTags probes are for research use only and are not validated for diagnostic or therapeutic purposes.