Promotors from myTXTL
myTXTL is a whole cell extract system relying on the endogenous TXTL machinery of E. coli. Hence, Sigma 70 transcription factor-specific and other E. coli promoters can directly be used to drive gene expression. Additionally, the standard T7 promoter system is compatible with myTXTL as well, when T7 RNA polymerase is present. For this, co-expression of T7 RNA polymerase from our helper plasmid pTXTL-P70a-T7rnap (e.g Cat# 502134) is recommended (this plasmid comes with the T7 Expression kits). Inducible promoters must have their inducer added and a higher DNA template concentration may be needed to maximize protein yield.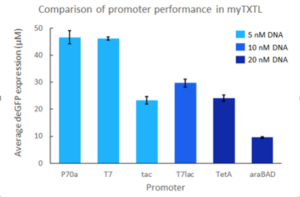
Propagation of P70-Plasmids
A particular E. coli strain is highly recommended for propagation of our P70-vectors. The strain KL740 cI857+ over-expresses a thermosensitive phage repressor protein and is crucial to maintain plasmid stability and integrity. Refer to Tech Note “Preparation of chemo-competent KL740 cells for amplification of P70a vectors” for further details.
Template Preparation
DNA template quality is critical for reproducible high-yield cell-free protein production.
o Plasmid: A 2-step plasmid purification procedure is highly recommended comprising 1) A standard plasmid preparation kit (e.g. Plasmid Plus Maxi Prep Kit from QIAGEN) followed by 2) A PCR clean-up procedure (e.g. PureLink from Invitrogen) with a final elution in nuclease-free water. Positive control plasmid(s) delivered with the myTXTL kits can serve as reference material to validate in-house plasmid purification procedures. Some myTXTL plasmids are available as ready-to-use, high-purity (HP) versions. For non-KL740 propagated plasmids, a 1-step purification with the ZymoPure plasmid prep kit is typically sufficient (P70a plasmids grown in KL740 cells require the extra purification step). Please note that plasmid sequences are available upon request.
o Linear DNA: For Linear DNA a simple PCR clean-up step as above is typically sufficient to maximize protein yield. Linear DNA from a manufacturer such as IDT or Twist can be used directly in the myTXTL reaction without further purification. See our IDT Technical Note for design recommendations here.
Optimizing TXTL reaction conditions
We recommend starting at 5 nM plasmid or 20 nM linear DNA for the template expressingnyour protein of interest. If further optimization is desired, the recommended concentration range for testing is:
o P70a Plasmid: 1-20 nM
o T7 Plasmid: 1-20 nM of T7 promoter plasmid in the presence of 0.1 nM pTXTL-P70a T7rnap. In some cases, lowering the concentration of the helper plasmid 2 to 4-fold improves the protein yield significantly
o Linear DNA: 5-30 nM
o Incubation
– Temperature: For most applications, myTXTL reactions perform best at 27C, but can also work well at 22-30C. Temperatures outside this range tend to be detrimental to protein yield. We strongly recommend using an incubator set at the desired temperature for best results.
– Vessel: The recommended incubation vessel for maximum protein yield is a 1.5/2mL reaction tube or a 96 well plate. Incubation in multi-well plates is excellent for highthroughput applications.
– Multi-well plate: For 96-well plates, we recommend the Costar Cat No. 3357 polypropylene plate with a Costar Storage Mat III Cat. No. 3080 cover.
Sample Downstream Processing
As a whole cell extract system, myTXTL reactions comprise a mixture of endogenous E. coli proteins in addition to your protein of interest once incubation is completed. Fusing a peptide tag (e.g. His-tag, Strep-tag, FLAG-tag, etc.) to your protein allows easy implementation of protein detection and/or purification techniques into your downstream processing. Note that MBP tags are not compatible with the myTXTL system. In many cases, protein purification prior to analysis is not required due to the open environment of the myTXTL system, see our Application Note with Arzeda for details here.

 Bluesky
Bluesky