Overview
myTXTL cell-free expression kits enable rapid protein expression, with many advantages over in vivo protein expression including the ability to deliver assayable protein in a single day and to skip time-consuming cloning and lysis/purification steps.
The myTXTL Pro Kit provides maximal expression of proteins without disulfide bonds, by leveraging the combined advantages of previous myTXTL kits.
Each myTXTL Pro Kit contains the Pro Master Mix, the Helper Plasmid (for T7 RNA polymerase expression), and the T7 deGFP Control Plasmid (for T7-promoter based expression of deGFP). To express proteins that contain disulfide bonds, please see our myTXTL Antibody/DS Kit.
- High yield – Produces enough protein for activity, screening, and biophysical assays (e.g. >1 mg/mL).
- Single day workflow– Enables quick turnaround times with an automation friendly procedure starting from linear or plasmid templates.
- Reduce processing – Skip lysis and purification to simplify protein expression, saving time and effort in downstream processing.
- No cloning required – The use of linear DNA eliminates the need for cloning, streamlining the expression process and saving 1-2 weeks versus in vivo expression.
- Express toxic proteins – The system supports the expression of toxic proteins, allowing researchers to study and characterize a wide range of target molecules with one kit.
- Maximum versatility – Supports constitutive and inducible E.coli and phage (T7) promoters, no unique plasmid system required.
Performance
myTXTL Pro Master Mix exceeds the performance of competitor systems
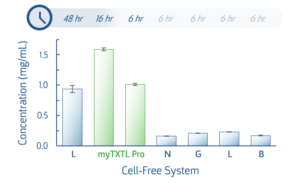
T7 deGFP expression was quantified following default expression protocols for myTXTL Pro Master Mix and several competitor systems. Reactions proceeded for the times indicated below.
Fig 1. Expression of the myTXTL T7 deGFP Control Plasmid was examined in multiple cell-free systems and quantified by deGFP fluorescence. Cell-free expression was conducted for the duration recommended for each system.
A wide range of proteins express at high-yield with myTXTL
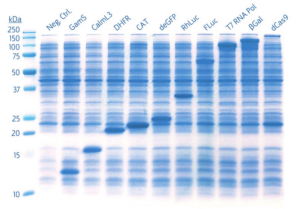
Proteins ranging from 12-160 kDa were expressed following the standard myTXTL protocol. 1 μL of each reaction was assayed by SDS-PAGE without purification.
Fig 2. SDS-PAGE visualization of proteins expressed in myTXTL Master Mix using5 nM Template DNA. 1 μL of myTXTL reaction is visualized per lane.
myTXTL Pro Master Mix expresses protein from common E.coli promoter systems
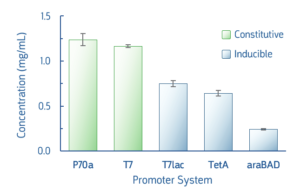
Several common promoter systems, both constitutive and inducible, were used to express deGFP in myTXTL Master Mix. Following 16-hour expression, yields of deGFP were ≥0.25 mg/mL for all tested promoters. The myTXTL system is highly flexible and able to use any promoter system used in E.coli protein expression, including T7 and pET vectors. The promoter strength and inducer used will affect protein expression level.
Fig 3. Comparison of deGFP expression following myTXTL expression. Reactions using T7lac, TetA, araBad promoters included inducer IPTG (1 mM), anhydrotetracycline(0.2 μg/mL), or arabinose (0.2 %), respectively.
Applications
myTXTL Pro Cell-Free Expression Kits are ideal for a range of research applications, including:
- High-throughput protein or enzyme engineering
- Protein synthesis (10-200kDa and higher)
- Production of soluble and membrane proteins
- Rebooting bacteriophages
- Prototyping of gene networks or gene circuits
- Construction of synthetic minimal or artificial cells
- Metabolic pathway optimization
- Metagenomic screening
- Teaching biological concepts
- And more!
Resources
Please note that some of our literature may refer to the myTXTL system using former product names.
FAQs
The myTXTL Pro Kit is intended for cell-free protein expression of proteins that do not require disulfide bonds. It also has the highest yield of our two kits. The Antibody/DS Kit is intended for the expression of proteins that do contain disulfide bonds such as antibodies and enzymes. The yield of the Antibody/DS kit varies with the protein expressed, but for deGFP it is about 30-50% the yield of the Pro Kit. This is still enough protein for most downstream applications.
For work involving proteins that require disulfide bond formation we recommend our myTXTL Antibody/DS Kit. It is possible that some proteins with just 1 disulfide bond, such as VHH/Nanobodies, may express and show activity in the myTXTL Pro Kit, but generally speaking disulfide bond containing proteins will have the highest yields and best activity in the myTXTL Antibody/DS Kit.
If you were using any of our 3 original myTXTL kits, those will be discontinued in August 2024. The myTXTL Pro Kit is now the kit that will serve your needs as it has all the properties of the old kits combined into one: it supports linear and plasmid DNA as well as all E. coli promoters and also enables T7 promoter-based expression through the addition of the Pro Helper Plasmid that expresses the T7 RNA polymerase. It’s a comprehensive solution for cell-free protein expression. All Pro Kits include the Pro Helper Plasmid and a T7 deGFP Positive Control Plasmid.
If you are interested in expressing proteins that contain disulfide bonds, the myTXTL Antibody/DS Kit is what you need.
Yes, myTXTL reactions for cell-free protein expression have been conducted from 2 to 100 uL volume, but above 50 uL we recommend shaking and/or switching to a reaction vessel with higher surface:volume ratio to allow proper oxygenation of the reaction mix. myTXTL reactions are very sensitive to the amount of dissolved oxygen. If using over 50 uL volume, a flat-bottomed ELISA, deep well plate or tissue culture plate may be advised along with shaking at 650 RPM. We advise testing such a setup with one of the positive control plasmids, such as T7 deGFP. The key is to balance oxygenation and avoid the reactions drying out due to too much surface area. Please refer to the appropriate kit manual for additional guidance on scaling up reaction volume.
As the myTXTL platform relies on the endogenous transcription and translation machinery of E. coli, a functional gene cassette must contain a promoter that can be transcribed by E. coli RNA polymerase and associated transcription factors (primarily Sigma 70) or by a T7/T3 RNA polymerase if those polymerases are expressed from a helper plasmid (available in our Toolkit). The ribosomal binding site should also be compatible with E. coli translation machinery. For more general advice on how to construct a functional gene cassette, please refer to the current myTXTL handbook.
myTXTL supports all promoters used in E. coli protein expression, including promoters that rely on the endogenous E. coli transcription machinery and those that require a separate RNA polymerase such as T7. All kits come with a Helper plasmid to express the T7 RNA polymerase and enable transcription from T7 promoters. If a promoter is used that is recognized by the E. coli RNA polymerase, then this helper plasmid is not needed. If using a plasmid with an inducible promoter, such as the pET vectors, you need to add your inducer IPTG at 1 mM to the reaction and you may want to explore plasmid concentration in the range of y your inducer like IPTG if it is an inducible promoter.
All myTXTL kits support plasmid or linear DNA templates as well as mRNA.
myTXTL supports all promoters used in E. coli protein expression, including promoters that rely on the endogenous E. coli transcription machinery and those that require a separate RNA polymerase such as T7. Inducible plasmids require that the inducer be added to obtain the highest protein yield. pET (T7lac promoter) systems need 1 mM IPTG, for example, otherwise follow the guidance in the following table for inducer and plasmid concentrations in the myTXTL reaction.
We recommend the following inducer and plasmid concentrations when using some common inducible promoters:
| Promoter | Inducer | Recommended Inducer Concentration in myTXTL | Units | Recommended Plasmid Template Concentration in myTXTL | Units |
| T7lac | IPTG | 1 | mM | 10 | nM |
| TetA | aTc | 20 | µg/mL | 20 | nM |
| araBAD | L-Arabinose | 2 | % | 20 | nM |
It may be necessary at times to add components to the myTXTL system from sources that contain material of unknown effect in the myTXTL system, the following is a guide for what chemicals/reagents may be tolerated without a loss in performance:
1) Glycerol is tolerated up to 0.1% of myTXTL reaction volume
2) DMSO is tolerated up to 1% of myTXTL reaction volume
3) EDTA is tolerated up to 0.1 mM of myTXTL reaction volume
4) Tris-HCl (pH 8) is tolerated up to 50 mM of myTXTL reaction volume
5) CaCl2 is tolerated up to 1 mM of myTXTL reaction volume
6) MgCl2 is tolerated up to 1 mM of myTXTL reaction volume
7) NaCl is tolerated up to 50 mM of myTXTL reaction volume
Yes, it is expected that protein yield resulting from linear templates in cell-free protein expression is diminished compared to its circular plasmid version. A decrease of 10-30% is considered to be within the normal range for the Pro kit. This decrease tends to be minimal in the Antibody/DS Kit.
myTXTL requires high quality template DNA, which should be free of nucleases (DNases, RNases) and inhibitors of the TXTL machinery (e.g. EDTA, ethidium bromide, SDS, Cl- ions, ethanol). Preparation of plasmid DNA with standard commercial kits usually involves sample treatment with RNase, which may not be completely removed during downstream processing. Thus, we strongly recommend subjecting the prepared DNA to either a commercial PCR clean-up kit or standard phenol-chloroform extraction and ethanol precipitation. Ideally, template DNA is in deionized, nuclease-free water. Please note, introducing Mg2+and K+ ions can compromise the kit performance, as they are extremely critical for transcription and translation, and are optimized in the master mix. Plasmid DNA prepared with a ZymoPure system purification kit or obtained from a commercial vendor can generally skip this extra purification step. Linear templates generated via PCR require just a single PCR Purification kit clean up (or magnetic bead cleanup) step or no clean-up if the PCR reaction is being diluted directly into the myTXTL reaction (be sure glycerol in the reaction is 0.1% or lower). See kit manual for additional recommendations.
Apart from standard biochemical methods like Coomassie-stained SDS-PAGE and western blot analysis, the great advantage of cell-free protein production is the open-system environment which allows the direct quantification and/or analysis of protein functionality in an activity assay without purification. Alternatively, for some activity assays downstream processing via affinity purification may be needed (if an affinity tag is present). If you choose SDS-PAGE analysis, you can either take a small sample (1-3 µL) directly from your TXTL reaction, or – to reduce background signal – precipitate proteins with TCA/acetone or ammonium-acetate/methanol following a standard protocol.
It is very important to avoid condensation of water on the lid of the reaction tube as it considerably increases the concentration of myTXTL reaction components and can lead to poor or irreproducible performance. Incubation in an incubator and water bath/Lab Armor beads is best. Water facilitates a faster heat transfer than air and a water bath shows low temperature fluctuation, which should – combined with a closed environment with constant temperature surrounding the entire tube – lead to higher reproducibility and yield. myTXTL reactions also work well on an Eppendorf ThermoMixer with a lid.
Yes! Parameters that influence protein production efficiency are:
- DNA design (promoter strength, position of affinity tag, TXTL elements)
- DNA purity
- DNA concentration
- Incubation temperature, time, and vessel
- Presence of folding helpers, chaperones, oxidizing agents
and should therefore be evaluated for optimization. Please also see our recommendations on these topics in the current myTXTL manual for your kit.
Consider if your recombinant protein requires co-factors like heavy metal ions or coenzymes to be functionally active. Those should be present during protein synthesis. Additionally, a low concentration of mild detergent (e.g. Triton-X-100, sodium dodecyl maltoside, or CHAPS) can be added to the reaction as well as molecular chaperones. Please note that the myTXTL system cannot introduce post-translational modifications like glycosylation or phosphorylation to your protein. Reducing the incubation temperature might help to prevent aggregation of the nascent polypeptide chain and to promote proper protein folding.
The myTXTL Pro Kit is best for rebooting bacteriophage and some modifications to the reaction mix may be needed for rebooting your phage, please refer to the manual for detailed suggestions on phage rebooting.
An additional application for the myTXTL Pro kit is that it can be used to reboot bacteriophage from a genome input. Several phages have been shown to replicate in the myTXTL system and they generally benefit from the following modifications:
1) The addition of 0.3 mM each of additional dNTPs (for replication of DNA genomes) and 0.5-4% PEG 8000 can be helpful for optimal production of phage. These components along with your genome, 9 uL of Pro Master Mix, and water if needed should add up to 12 uL and be assembled in a 1.5/2 mL tube. Larger reaction volumes may require shaking as described in the manual for larger scale reactions. Incubate overnight as in our protocol.
2) You may also need to optimize the phage genome concentration if it is not already published. 0.25 nM has worked well for T7 but T4 was optimal at 1 nM. In part this is dependent on the quality of the genome. It is extremely important that the phage genome is of very high purity, free of contaminants and of high integrity.
3) Commercially available T7 phage genomic DNA can be used directly in the Pro Master Mix as a positive control (we have used DNA from BocaScientific: https://bocascientific.com/310000-86-detail).
Several factors can hinder protein synthesis in the myTXTL cell-free protein expression system, including:
- Master mix inactivation due to improper storage. The myTXTL master mix must be stored at -80°C and number of freeze-thaw cycles should be minimized.
- Improper reaction setup, Please review the recommendations to set up a myTXTL reaction in the current myTXTL manual.
- Contamination of myTXTL reaction with nuclease. To avoid nuclease contamination, wear gloves and use nuclease-free water, sterilized tips and tubes. Use deionized, nuclease-free, "molecular grade" water for myTXTL reaction setup.
Batch-to-batch variation can cause varying levels of TXTL inhibitor contamination present in the plasmid solution. Please follow our recommendations on how to prepare template plasmid DNA for TXTL reactions in the current myTXTL manual.
PCR products can be directly used as DNA template for cell-free expression so long as the final glycerol concentration in the reaction is 0.1% or lower and the PCR reactions on average are making enough linear DNA to facilitate the protein yield needed. If a known fixed template concentration is preferred, PCR products should be subjected to a standard PCR clean up procedure with a final elution in molecular biology grade, nuclease-free water. This might also slightly boost the protein yield per reaction. Linear DNA can also be ordered and used directly in myTXTL from several commercial DNA suppliers. Please refer to the applicable myTXTL manual and our Application Note with IDT for information about linear DNA template design requirements, or reach out to techsupport@arbor.daicel.com with questions about specific sequence recommendations.
Unfortunately, this may lead to considerably decreased performance or even loss of function. To ensure highest kit performance, make sure to store the myTXTL cell-free protein expression kit master mix at -80 °C and freeze as soon as possible after usage.
We share plasmid and linear DNA sequences that have been tested to work well in myTXTL, including kit plasmid controls, to allow users to optimize their designs with respect to 5' and 3' sequences that enable optimal expression for their application. Please contact techsupport@arbor.daicel.com with any questions.
Yes. The myTXTL master mix contains tRNAs for seven codons rarely used in E. coli to enable expression of eukaryotic proteins.
Using linear DNA templates greatly increases the speed of the design-build-test-learn cycle as laborious steps like cloning, transformation and purification are no longer necessary. This is particularly useful when working with a high number of variants of a single protein that need to be studied and validated. The reduced costs of linear DNA can also expand the sampled sequence space for protein designs compared to the plasmid format.
deGFP is a N- and C-terminally truncated version of the reporter eGFP that is more translatable in cell-free systems. The excitation and emission spectra as well as fluorescence properties of deGFP and eGFP are identical which enables the use of commercial eGFP protein to be used in a standard curve to quantify the deGFP in the reaction.
Yes. Due to the manufacturing process, there might be a small pellet visible. It is critical that you resuspend this pellet back into the myTXTL Master Mix completely before aliquoting it to set up your myTXTL reaction(s).
Sample handling and storage is mainly determined by the stability of your molecule of interest (protein, DNA, RNA) and thus optimal conditions may need to be evaluated. But to ensure sample integrity, we would recommend to either process the myTXTL reaction immediately after performing the incubation or store it at ≤ -20 °C.
Limit the number of freeze-thaw-cycles as much as possible. Our studies have indicated that up to five freeze-thaw-cycles do not negatively influence protein production efficiency of the myTXTL Master Mix when using flash freezing in liquid nitrogen and subsequent -80C storage.
Most importantly, the excitation and emission wavelength should match the fluorescence properties of deGFP/eGFP (e.g. λEm 488 nm, λEx 535 nm). Other reader settings such as reading mode, integration time and gain value should be chosen under consideration of high well-to-well fluorescence reading reproducibility.


 Bluesky
Bluesky