myTXTL cell-free protein expression enables development of sensors that detect important biomarkers including proteins and nucleic acids. In such sensors, molecular binding events lead to amplification of a reporter signal that can be fluorescent, colorimetric, luminescent, or electrochemical. Through the use of DNA or RNA that bind to molecules of interest, cell-free protein expression can be linked to successful binding events. Micro-RNAs (miRNAs) as biomarkers for disease have seen increasing interest as targets for bedside diagnostics.
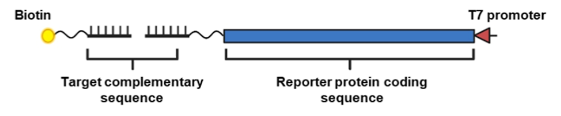
In a recent paper, Y-H. Min et al. showcased how cell-free protein expression can be used to quantify concentrations of disease-related miRNA in the Journal ACS Applied Bio Materials1. The researchers designed split DNA probes (Figure 1) with 2 probes for any target miRNA sequence. One DNA probe is conjugated to biotin for purification while the other probe contains a reporter protein sequence such as green fluorescent protein (GFP) or luciferase. The miRNA target binds both the biotin conjugated probe and the probe with the reporter protein. Without miRNA bridging the split probe, no reporter sequence template can be bound.
Figure 1: Split-probe design
Once miRNA binds to both probes, streptavidin-conjugated magnetic beads can then pull down the entire complex, including the reporter protein DNA template. A cell-free protein expression mix, like myTXTL, is then added to the bead-bound DNA to begin expression of fluorescent or luminescent reporter proteins. Y-H. Min et al. demonstrate the ability to monitor both single and dual miRNA targets with high specificity and sensitivity. For dual miRNA targets, split-probes encoding a different fluorescent reporter protein for each miRNA were used simultaneously. With a luciferase as the reporter protein, miRNAs were quantified at concentrations as low as 0.5 fM!
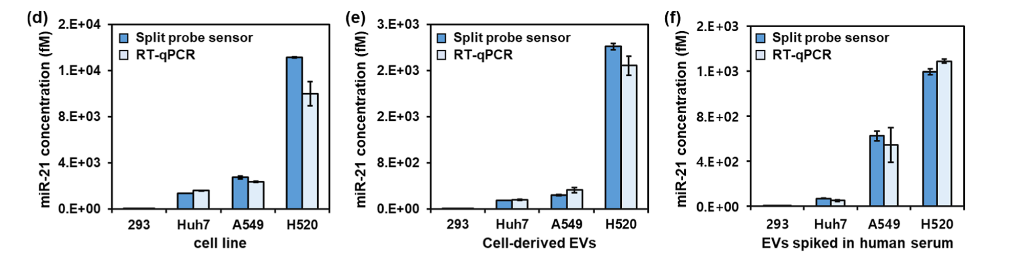
After the proof-of-concept experiments, Y-H. Min et al. compared the split-probe sensor with a luciferase protein reporter to standard quantitative real-time PCR (RT-qPCR) for miRNA miR-21 detection in clinically relevant samples. Three human cancer cell lines and extracellular vesicles (EVs) derived from cancer cell lines were shown to upregulate miR-21 expression relative to a HEK293 control (Figure 4d-e). Further, miRNA in EVs spiked into human serum to mimic clinical samples were quantified using the split-probe sensor and standard RT-qPCR with comparable results, indicating the split-probe method is highly accurate (Figure 4f). This study demonstrates the power of cell-free protein expression to sense biomarkers, giving specific and sensitive diagnostic output at low cost.
Figure 4d-f: Split-probe sensor clinical sample performance compared to RT-qPCR. miR-21 concentrations are determined from three sources: cell lines (d), cell-derived EVs (e), and EVs added to human serum.
Only binding-activated DNA or RNA probes linked to a protein reporter template DNA are required to harness the power of myTXTL for biosensor signal amplification. Once activated DNA or RNA probes and the protein reporter template are added to myTXTL Master Mix signal amplification will begin (Protein Expression). Dive into the paper below to learn more!
1 Min, Y-H., Hong, Y., Kim, C-H., et. al. Split Probe-Induced Protein Translational Amplification for Nucleic Acid Detection. ACS Applied Bio Materials (2024). https://doi.org/10.1021/acsabm.4c01187



 Bluesky
Bluesky