Overview
myTXTL Cell-free Expression Kits offer an efficient, easy-to-use solution for protein synthesis, enzyme engineering and synthetic biology in industrial and academic applications. They are based on our myTXTL platform, a well-characterized technique that has been employed in synthetic biology labs globally for a wide variety of applications including synthesis of toxic proteins, rapid enzyme engineering, and bacteriophage production.
myTXTL Cell-free Expression Kits pair well with automated liquid handling systems, making them ideal for rapid design-build-test cycles when screening libraries or analyzing genetic parts and gene circuits.
Key benefits
- Fast processing—Save time by avoiding transformation, clone selection and cell lysis, enabling rapid design-build-test cycles.
- High controllability—Easily adjust experimental parameters to meet your project requirements.
- User-friendly—Safe, GMO-free handling.
- Multiple size options—Bulk and OEM options are available.
Product selector
To help select the correct myTXTL Cell-free Expression Kit for your project, consult the table below.
| Purpose | Product |
|---|---|
| myTXTL Sigma 70 Master Mix Kit Learn More |
| myTXTL Linear DNA Expression Kit Learn More |
| myTXTL T7 Expression Kit Learn More |
Resources
FAQs
As the myTXTL platform relies on the endogenous transcription and translation machinery of E. coli, a functional gene cassette must contain a promoter that can be transcribed by E. coli RNA polymerase and associated transcription factors (primarily Sigma 70) or by a T7/T3 RNA polymerase if those polymerases are expressed from a helper plasmid (available in our Toolkit). The ribosomal binding site should also be compatible with E. coli translation machinery. For more general advice on how to construct a functional gene cassette, please refer to the current myTXTL handbook.
We share all DNA plasmid and linear fragment sequences in our portfolio to allow users to optimize their designs with respect to 5' and 3' sequences that enable optimal expression for their application, ranging from single protein expression to complex gene circuits and metabolic pathways. Plasmid or linear DNA can be ordered online from various DNA synthesis companies.
Yes! Parameters that influence protein production efficiency are:
- Gene cassette construction (promoter strength, position of affinity tag, TXTL elements)
- DNA purity
- DNA concentration
- Incubation temperature, time, and vessel
- Presence of folding helpers, chaperones, oxidizing agents
and should therefore be evaluated for optimization. Please also see our recommendations on Template Design in the current myTXTL handbook.
Yes. The myTXTL master mix contains tRNAs for seven codons rarely used in E. coli to enable expression of eukaryotic proteins.
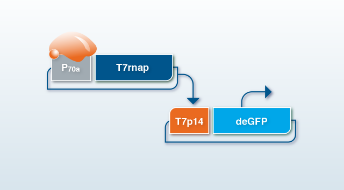
Yes! That only requires the addition of the plasmid coding for T7 RNA polymerase under transcriptional control of a σ70-specific promoter, e.g. P70a-T7rnap and possibly your inducer like IPTG if it is an inducible promoter. The optimum concentration of P70a-T7rnap is usually between 0.1 nM and 1 nM. Higher concentration normally does not increase protein yield. The more important parameter for efficient protein expression is the concentration of the plasmid that encodes for your protein of interest downstream of the T7 promoter, which will be most likely in the range of 5-20 nM.
Apart from standard biochemical methods like Coomassie-stained SDS-PAGE and western blot analysis, the great advantage of cell-free protein production is the open-system environment which allows the direct quantification and/or analysis of protein functionality in an activity assay without purification. Alternatively, for some activity assays downstream processing via affinity purification may be needed (if an affinity tag is present). If you choose SDS-PAGE analysis, you can either take a small sample (1-3 µL) directly from your TXTL reaction, or – to reduce background signal – precipitate proteins with TCA/acetone or ammonium-acetate/methanol following a standard protocol.
Utilizing linear DNA templates greatly increases the speed of the design-build-test-learn cycle as laborious steps like cloning, transformation and purification are no longer necessary. This is particularly useful when working with a high number of variants of a single protein that need to be studied and validated. The reduced costs of linear DNA can also expand the sampled sequence space for protein designs compared to the plasmid format.
deGFP is a N- and C-terminally truncated version of the reporter eGFP that is more translatable in cell-free systems. The excitation and emission spectra as well as fluorescence properties of deGFP and eGFP are identical which enables the use of commercial eGFP protein to be used in a standard curve to quantify the deGFP in the reaction.
The myTXTL Linear DNA Expression Kit is based off our myTXTL Sigma 70 Master Mix Kit, but has been further engineered to efficiently produce soluble and membrane proteins using linear DNA templates without the need for nuclease inhibitors like GamS. Simply add linear DNA template to the optimized master mix to begin protein synthesis.
Efficient in vitro protein production is highly dependent on the quality of the template DNA, which should be free of nucleases (DNases, RNases) and inhibitors of the TXTL machinery (e.g. EDTA, ethidium bromide, SDS, Cl- ions, ethanol). Preparation of plasmid DNA with standard commercial kits usually involves sample treatment with RNase, which may not be completely removed during downstream processing. Thus, we strongly recommend subjecting the prepared DNA to either a commercial PCR clean-up kit or standard phenol-chloroform extraction and ethanol precipitation. Ideally, template DNA is suspended in nuclease-free water. Please note, introducing Mg2+and K+ ions can compromise the kit performance, as they are extremely critical for transcription and translation, and are optimized in the master mix.