Overview
Leveraging our expertise in designing and manufacturing high-performance NGS enrichment panels for pathogens, Daicel Arbor Biosciences rapidly developed this panel for complete, targeted SARS-CoV-2 viral genome sequencing early in the COVID pandemic, in response to urgent requests from the research community. This hybridization capture kit enables researchers studying epidemiology, phylogenetics, and viral dynamics of SARS-CoV-2.
The myBaits Expert Virus SARS-CoV-2 panel is based on multiple full and partial genome sequences available in the NCBI database. The flexible nature of hybridization capture allows these probes to capture novel variants, including point mutations, small or large indels, or other genomic changes. The design of the probes is compatible with enriching both long and short NGS library molecules, even from degraded targets.
- Convenient–Use the same NGS library preparation or protocol for shotgun sequencing.
- Open platform–Compatible with any NGS library platform, including long-read sequencing.
- Complete solution–Kits include all necessary reagents to perform enrichment of NGS libraries.
Want data even faster? Leverage our expert myReads® NGS services.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Performance
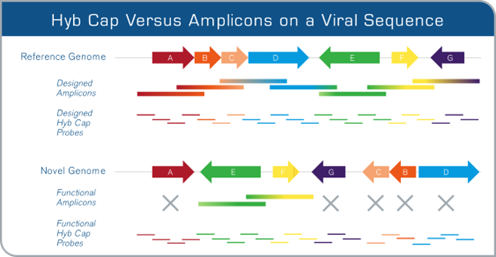
Hybridization capture tolerates both rearrangements and sequence variation
Both hybridization capture and amplicon sequencing can effectively enrich for known sequence regions. However, only hybridization capture can retrieve target sequences that have significant rearrangements and/or mutations relative to the reference used for probe design, for example when capturing genomic content from novel microbial genomes.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Resources
Application Note
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.