Overview
Recovering appreciable full nuclear genome coverage from metagenomic, environmental, or ancient DNA samples with shotgun sequencing data is expensive and bioinformatically intense. mBaits Expert WGE kits from Daicel Arbor Biosciences help overcome these challenges.
Our WGE technology can cost-effectively produce enrichment baits representative of an entire nuclear genome. This enables capture of genome-wide endogenous DNA from complex metagenomic samples, such as environmental or ancient DNA. Need custom WGE baits? All that is required is a sample of high-quality genomic DNA from an organism of interest, or a close relative, even if the genome has not been sequenced.
Custom or predesigned bait sets – Choose from our in-stock kits for ancient DNA or select from an organism of interest for a custom-manufactured set.
Efficient genome-wide sequencing – Isolate a genome of interest from complex samples.
Perfect for exploratory studies – Sequenced genome is not required.
Want data even faster? Our expert myReads NGS services can process many degraded or ancient DNA samples to help move your project forward.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Available Panels
Daicel Arbor Biosciences has multiple genomic DNA (gDNA) WGE panel options for human (Caucasian, African, or Japanese Male) which are in stock and ready for quick shipment.
We can also produce Custom WGE panels for any organism. We require a high-purity, high-molecular weight gDNA sample to be (user-supplied or sourced from a commercial provider) shipped to our manufacturing facility in Ann Arbor, MI, USA. Using our technology we transform the gDNA into ready-to-use, genome-wide target capture baits. Custom WGE kits typically require 2-4 weeks for production (starting from when we receive the suitable genomic DNA sample). For more information about this option contact us.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Applications
Due to the high sequencing depth required to observe appreciable coverage of whole-genome data, especially from degraded or complex samples, Whole Genome Enrichment (WGE) kits are generally best suited for exploratory research projects. If the research goal is to recover specific genes or markers of interest, then a focused myBaits Custom panel is more appropriate for maximizing sequencing efficiency. Our experts can help design the Custom panel from your target regions of interest.
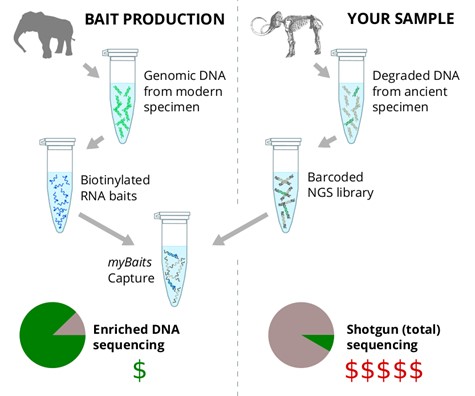
Comparison of myBaits WGE kit with traditional shotgun techniques
Baits can be produced from any pure eukaryotic or prokaryotic genomic DNA source; modern elephant, for example. Daicel Arbor Biosciences’ unique process converts this gDNA into a pool of biotinylated RNA baits. These baits can then enrich corresponding molecules from a user-supplied NGS library–for example, from an ancient mammoth–via the process of myBaits in-solution hybridization capture. This enriched library allows for NGS that is orders-of-magnitude more efficient than is possible using the full, non-enriched library.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Resources
Protocol Note: Please see tips in the manual PDF as well as FAQ tab for recommended procedure modifications for performing WGE from metagenomic/ancient DNA samples. In the v5 manual, following the “High Sensitivity” protocol is recommended.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
FAQs
To produce a Custom WGE panel, a high-purity, high-molecular weight genomic DNA (gDNA) sample is required to be shipped to Arbor’s manufacturing facility in Ann Arbor, MI, USA.
If you do not have access to the appropriate gDNA, for some organisms Arbor may be able to provide the additional service of acquiring the gDNA sample(s) from a third-party vendor.
Custom gDNA sourcing service is only upon prior arrangement and approval by Arbor. This service is only available for species whose genomic DNAs are available from vendors supplying the US. The gDNA sample(s) must be available for Arbor to purchase in sufficient quantity as a commercial entity without any special licenses, acquisition or use permits, or memberships.
Please contact us to provide specific details on the gDNA source(s)/species that you are interested in, and we will review the request to determine feasibility and pricing.
Target capture necessarily requires subjecting your libraries to a bottleneck, wherein target molecules are captured and enriched, and non-target molecules are therefore removed. To have sufficient unique molecules for good sequencing coverage of your targets, successful captures DEPEND on the input of sufficiently complex libraries. For this reason, for libraries with a significant non-target component (e.g., ancient, forensic, or environmental samples), and especially for WGE captures with a very large full nuclear genome target size, we strongly recommend maximizing the target component in each capture by using as much input library as possible (up to 2 µg+), and consider two rounds of capture for higher percentage of reads on-target.
For best results, it is recommended that only amplified (non-PCR-free) NGS libraries are used for target capture. This provides multiple copies of each starting template molecule, increasing the chance of each individual molecule getting enriched. However if you need more starting material to reach the recommended amount, it is generally preferable to generate more library from fresh genomic DNA or a new batch of indexed library, rather than through extra amplification. This is because while some amplification is good, over-amplification risks reducing the observable complexity of your libraries through the uneven action of PCR bias, as some molecules will become relatively more abundant while others become rare. This is also true for manipulating your libraries after capture: amplify your post-capture libraries the minimum number of cycles necessary to reach the molarity required by your sequencing facility.
We generally do NOT recommend pooling multiple samples per capture reaction for very degraded and/or rare targets (e.g. ancient DNA), or for very large targets (e.g. a WGE baitset targeting a full nuclear genome). In fact, for highly degraded samples, you may achieve better results by performing multiple parallel WGE reactions per sample.
The myBaits WGE product line ONLY offers bait production directly from high quality genomic DNA precursor material. This gDNA must be made physically available to us in order to manufacture a myBaits WGE Custom kit (either extracted in your lab or sourced from a third-party gDNA supplier).
Daicel Arbor Biosciences does have the ability to design and synthesize synthetic customized bait oligo pools via our standard myBaits Custom DNA-Seq, RNA-Seq, and Methyl-Seq products. However the myBaits Custom synthetic oligo approach is generally not an option for targeting entire large nuclear genomes (e.g. from eukaryotic organisms such as plants or animals) due to prohibitively large numbers of baits that would be required. However if you are working with an organism that has a much smaller genome (e.g. one or more bacteria), or are interested in capturing only a portion of a much larger genome, then a myBaits Custom kit is likely the most effective option.
Please contact us with details about your project goals and budget, so we can discuss project options.
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.
Publications
Proceedings of the National Academy of Sciences | January 17, 2023
Science | July 6, 2018
Proceedings of the National Academy of Sciences | April 18, 2017
Microbial Ecology | April 1, 2017
Whole-Genome Enrichment Provides Deep Insights into Vibrio cholerae Metagenome from an African River
Molecular Biology and Evolution | May 1, 2014
Ancient Whole Genome Enrichment Using Baits Built from Modern DNA
myBaits kits are for research use only and are not validated for diagnostic or therapeutic purposes.


 Bluesky
Bluesky