Yes, myTXTL Linear DNA Expression Kit allows the use of both, linear and circular/plasmid templates.
Sample handling and storage is mainly determined by the stability of your molecule of interest (protein, DNA, RNA) and thus optimal conditions may need to be evaluated. But to ensure sample integrity, we would recommend to either process the myTXTL reaction immediately after performing the incubation or store it at ≤ -20 °C.
Limit the number of freeze-thaw-cycles as much as possible. Our studies have indicated that up to five freeze-thaw-cycles do not negatively influence protein production efficiency of the myTXTL Master Mix when using flash freezing in liquid nitrogen and subsequent -80C storage.
Yes, please see examples in the Publications section; filter for myTXTL. Please note, that every gene circuit should start with a σ70-specific promoter like P70a.
Most importantly, the excitation and emission wavelength should match the fluorescence properties of deGFP/eGFP (e.g. λEm 488 nm, λEx 535 nm). Other reader settings such as reading mode, integration time and gain value should be chosen under consideration of high well-to-well fluorescence reading reproducibility.
Yes. For all plasmids containing the lambda phage promoter (P70a, P70b, P70c, P70d) it is extremely crucial to use E. coli KL740 as the transformation strain. When cultivated below 30°C, this strain over-expresses the lambda phage repressor protein Cl857 that represses P70 promoters, thus ensuring high transformation efficiency and plasmid stability. KL740 can be purchased from E. coli Genetic Stock Center (Yale) [CGSC#: 4382] or from Daicel Arbor. For all other plasmids, a standard laboratory E. coli cloning strain like JM109 or DH5alpha is sufficient.
Yes, Daicel Arbor Biosciences has an NGS services division adept at preparing and working with ancient DNA libraries. Learn more about our myReads NGS laboratory and sequencing service options, and contact us today to discuss your NGS project goals with our team of ancient DNA experts.
Yes. Specific recommendations for library pooling for co-enrichment with myBaits Custom Methyl-Seq kits can be found in the applicable myBaits manual.
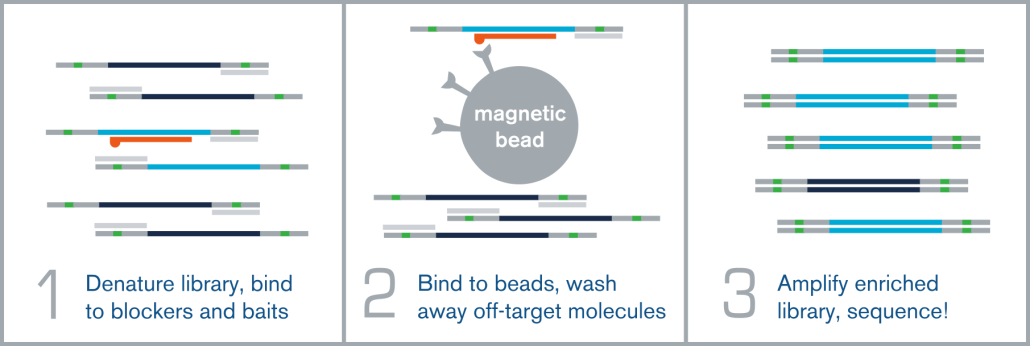
Hybridization capture is integrated into the overall NGS workflow immediately before sequencing on an NGS platform, such as Illumina. A fully sequenceable, barcoded/indexed NGS library is made from bisulfite or enzymatically-converted DNA. A pool of these multiple barcoded NGS libraries is then denatured, and allowed to anneal to complementary target-specific biotinylated probes/baits. These bait:library complexes are then bound to streptavidin-coated magnetic beads via the biotin on the probes, which are washed to remove non-specifically bound molecules. The remaining “enriched” library molecules are then released from the baits and amplified before sequencing.
Note! You may know the “hybridization capture” technique by another name, such as:
- Target enrichment
- Target capture
- Probe capture
- Exon capture
- Capture sequencing / sequence capture
- Hybridization sequencing / hyb-seq
Use myBaits Custom Methyl-Seq with PCR-amplified and amplifiable NGS libraries generated from bisulfite- or enzymatically-converted nucleic acids in which non-methylated cytosines have been converted to uracils, and following PCR amplification these positions are now thymines. Compatible formats include Illumina® TruSeq®-style, Illumina Nextera® Flex-style, Ion Torrent®, or other libraries with universal adapter priming sites. Do NOT use myBaits with PCR-free libraries; additionally, myBaits are incompatible with libraries made using original Nextera or Nextera XT library preparation kits, or any library type containing biotin. Dual-indexed libraries are strongly recommended to reduce the hazard of mis-indexing induced by PCR jumping events. The applicable myBaits manual provides detailed protocol instructions for enriching libraries intended for methylation sequencing.
If you are using a never-before-tried library prep protocol to pair with your myBaits kit, we recommend that you first perform some total library (shotgun) sequencing before doing myBaits enrichment. This is important in order to verify that your chosen library prep protocol/kit generates libraries of sufficient complexity and minimal bias in your hands, otherwise you may experience poor target capture results. High quality libraries are absolutely essential for achieving a successful target capture project.
Use myBaits with PCR-amplified and amplifiable NGS libraries, including Illumina TruSeq® -style, Illumina Nextera® Flex-style, Ion Torrent, or other libraries with universal adapter priming sites. It is NOT recommended to use myBaits with PCR-free libraries. Additionally, myBaits are incompatible with libraries made using original Nextera or Nextera XT library preparation kits, or any library type containing biotin. Dual-indexed libraries are strongly recommended to reduce the hazard of mis-indexing induced by PCR jumping events. The applicable myBaits manual provides detailed protocol instructions for enriching libraries for sequencing on short- and/or long-read platforms (e.g. PacBio® or Oxford Nanopore Technologies®).
If you are using a never-before-tried library prep protocol to pair with your myBaits kit, we recommend that you first perform some total library (shotgun) sequencing before doing myBaits enrichment. This is important in order to verify that your chosen library prep protocol/kit generates libraries of sufficient complexity and minimal bias in your hands, otherwise you may experience poor target capture results. High quality libraries are absolutely essential for achieving a successful target capture project.
Provided below are a list of companies that sell NGS library prep kits that are known to be compatible with myBaits. This is NOT an exhaustive list; there are many other unlisted options that are also compatible with myBaits. Also, kits on this list may not necessarily be appropriate for your samples. NGS library prep is not “one size fits all”; different factors such as sample type, DNA input amount, genome complexity, and sequence composition may influence the type of library prep kit that would be best for your application. For example, low input, degraded, and/or damaged DNA templates may require special handling (see below) and/or modifications to commercial kits.
Contact these and other manufacturers to learn about your options and find what works best for your samples and project needs:
- Biosearch / Lucigen
- Claret Bioscience
- Illumina
- New England Biolabs
- Kapa Biosystems
- PerkinElmer / Bioo Scientific
- Rubicon Genomics / Takara
- Swift Biosciences / IDT
To produce a Custom WGE panel, a high-purity, high-molecular weight genomic DNA (gDNA) sample is required to be shipped to Arbor’s manufacturing facility in Ann Arbor, MI, USA.
If you do not have access to the appropriate gDNA, for some organisms Arbor may be able to provide the additional service of acquiring the gDNA sample(s) from a third-party vendor.
Custom gDNA sourcing service is only upon prior arrangement and approval by Arbor. This service is only available for species whose genomic DNAs are available from vendors supplying the US. The gDNA sample(s) must be available for Arbor to purchase in sufficient quantity as a commercial entity without any special licenses, acquisition or use permits, or memberships.
Please contact us to provide specific details on the gDNA source(s)/species that you are interested in, and we will review the request to determine feasibility and pricing.
Target capture necessarily requires subjecting your libraries to a bottleneck, wherein target molecules are captured and enriched, and non-target molecules are therefore removed. To have sufficient unique molecules for good sequencing coverage of your targets, successful captures DEPEND on the input of sufficiently complex libraries. For this reason, for libraries with a significant non-target component (e.g., ancient, forensic, or environmental samples), and especially for WGE captures with a very large full nuclear genome target size, we strongly recommend maximizing the target component in each capture by using as much input library as possible (up to 2 µg+), and consider two rounds of capture for higher percentage of reads on-target.
For best results, it is recommended that only amplified (non-PCR-free) NGS libraries are used for target capture. This provides multiple copies of each starting template molecule, increasing the chance of each individual molecule getting enriched. However if you need more starting material to reach the recommended amount, it is generally preferable to generate more library from fresh genomic DNA or a new batch of indexed library, rather than through extra amplification. This is because while some amplification is good, over-amplification risks reducing the observable complexity of your libraries through the uneven action of PCR bias, as some molecules will become relatively more abundant while others become rare. This is also true for manipulating your libraries after capture: amplify your post-capture libraries the minimum number of cycles necessary to reach the molarity required by your sequencing facility.
We generally do NOT recommend pooling multiple samples per capture reaction for very degraded and/or rare targets (e.g. ancient DNA), or for very large targets (e.g. a WGE baitset targeting a full nuclear genome). In fact, for highly degraded samples, you may achieve better results by performing multiple parallel WGE reactions per sample.
The myBaits WGE product line ONLY offers bait production directly from high quality genomic DNA precursor material. This gDNA must be made physically available to us in order to manufacture a myBaits WGE Custom kit (either extracted in your lab or sourced from a third-party gDNA supplier).
Daicel Arbor Biosciences does have the ability to design and synthesize synthetic customized bait oligo pools via our standard myBaits Custom DNA-Seq, RNA-Seq, and Methyl-Seq products. However the myBaits Custom synthetic oligo approach is generally not an option for targeting entire large nuclear genomes (e.g. from eukaryotic organisms such as plants or animals) due to prohibitively large numbers of baits that would be required. However if you are working with an organism that has a much smaller genome (e.g. one or more bacteria), or are interested in capturing only a portion of a much larger genome, then a myBaits Custom kit is likely the most effective option.
Please contact us with details about your project goals and budget, so we can discuss project options.
We offer complementary myTags Custom (F)ISH probe bioinformatic design service using our proprietary design algorithms for most types of (F)ISH projects. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will apply our advanced proprietary probe design algorithms to craft a custom probe design for your targeting needs, for any organism or application. Our scientific team has decades of experience designing custom probes for a wide variety of applications, including multi- or single locus/gene localization, chromosome painting, chromosomal indexing/barcoding, haplotyping, and more.
Only the highest specificity regions with minimal background noise and cross-reactivity for premium performance in any downstream experimental application would be selected for the probe design.
And with our transparent and straightforward scientific communication process, you always have full control and ownership over the oligo sequences for each and every myTags Custom probeset.
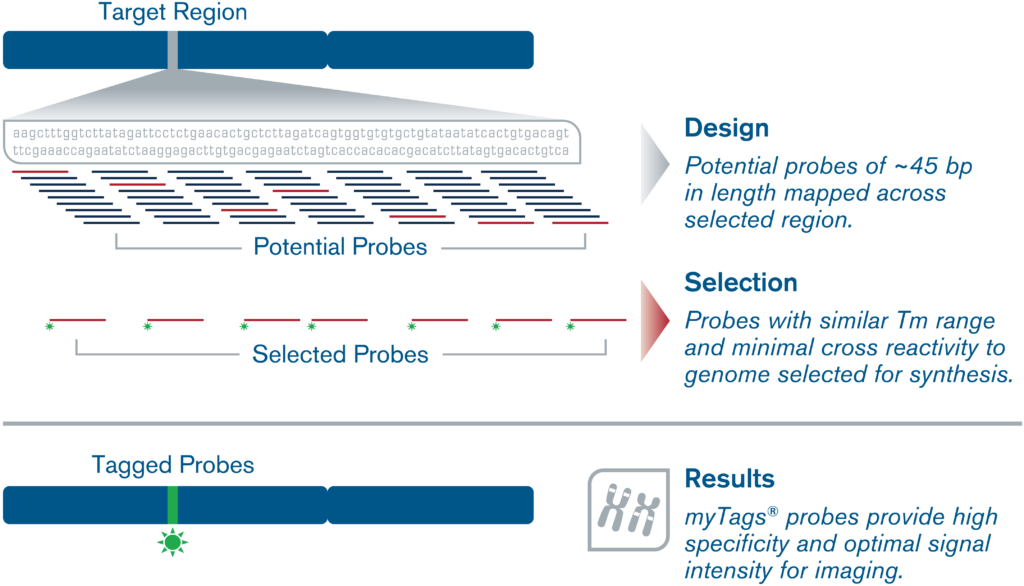
Generally we recommend probe densities between 3-10 probes per kilobase for target regions larger than 50kb. For target regions between 10-50kb, probe densities should be on the higher end of that range, and we may recommend using multiple fluorophores per probe to boost the signal.
We can work with any sequence to design probes. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will advise you on the most appropriate synthesis configuration for your desired project needs, which depends on the number of individual probe sequences required for your design. This is primarily driven by size of your target region(s), but also your experimental setup and goals.
You can choose to receive your individual probe pool(s) either as non-labeled (a.k.a. “immortal”) amplifiable substrate(s) ready for labeling in your lab, or receive as labeled, ready-to-use probe pool(s) for your experiments.
Regardless, all unique probesets are delivered individually, and include individual final probeset pool composition verification via next-generation sequencing.
myBaits hybridization capture kits include:
- Biotinylated RNA probes, with sequences corresponding to your custom design or a predesigned catalog option
- Hybridization and wash reagents
- For myBaits Custom kits, optional custom probe design informatics service (= expert bioinformaticians design and filter bait sequences and provide summary report and recommendations)
You will receive enough probes and reagents for performing the stated number of individual capture reactions of your kit size (e.g., 16 reactions) according to our current protocol. Please note that there are some additional reagents and equipment you will need to supply in order to perform a myBaits capture. Please review the list of required materials in the applicable myBaits manual to make sure you have everything you need before starting your experiments.
We also offer reagents for preparing libraries from DNA samples in advance of performing the myBaits hybridization capture step. Please visit Library Prep Kit for myBaits for more information.
If you are looking to outsource your project to a full-service laboratory and bioinformatics services group, please visit our myReads NGS laboratory and bioinformatics services page for more information about our comprehensive targeted sequencing service options (library preparation, target capture, next-generation sequencing, and optional analysis).
In the context of hybridization capture for targeted next generation sequencing, we use the terms interchangeably. Some fields prefer one term over the other, so we use both terms.
Turnaround time for myBaits targeted next generation sequencing kits varies based on design.
For new myBaits Custom baitset designs, the estimated manufacturing lead time is ~3-4 weeks minimum, starting from when your order is received and you have approved the final design. In addition, please consider that if you utilize our included bait design services, we will typically be in correspondence for an additional upfront period (up to several weeks) regarding a design before manufacturing can begin. Please also remember to accommodate any additional time for your collaborators to approve the final design, if applicable.
For myBaits Expert (catalog) kits or reorders of myBaits Custom kits with designs previously manufactured by Daicel Arbor Biosciences, the estimated manufacturing lead time is up to ~1-2 weeks from the time an order is received.
All myBaits kits include a specific protocol for their use as well as almost all of the reagents required to deploy them. In the manual, you will find the complete list of required supplies (reagents and equipment) that you will need in order to perform the captures.
Please see the applicable myBaits manual for detailed protocol instructions for enriching from Standard, High-Sensitivity, Long-Insert, or other specialty target/sample types.

Hybridization capture is integrated into the overall next generation sequencing workflow immediately before sequencing on an NGS platform, such as Illumina. A fully sequenceable, barcoded/indexed NGS library (or pool of multiple libraries) is denatured, and allowed to anneal to complementary target-specific biotinylated probes/baits. These bait:library complexes are then bound to streptavidin-coated magnetic beads via the biotin on the probes, which are washed to remove non-specifically bound molecules. The remaining “enriched” library molecules are then released from the baits and amplified before sequencing.
Note! You may know the “hybridization capture” technique by another name, such as:
- Target enrichment
- Target capture
- Probe capture
- Exon capture
- Capture sequencing / sequence capture
- Hybridization sequencing / hyb-seq
- Hybridization capture / hyb-cap
Specific recommendations for per-library input mass for different enrichment project types can be found in the applicable myBaits manual.
Target capture necessarily requires subjecting your libraries to a bottleneck, wherein target molecules are captured and therefore enriched, and non-target molecules are therefore removed. To have sufficient unique molecules for good sequencing coverage of your targets, successful captures DEPEND on the input of sufficiently complex libraries.
For best results, it is recommended that only amplified (non-PCR-free) NGS libraries are used for target capture. This provides multiple copies of each starting template molecule, increasing the chance of each individual molecule getting enriched. However if you need more starting material to reach the recommended amount, it is generally preferable to generate more library from fresh genomic DNA or a new batch of indexed library, rather than through extra amplification. This is because while some amplification is good, over-amplification risks reducing the observable complexity of your libraries through the uneven action of PCR bias, as some molecules will become relatively more abundant while others become rare. This is also true for manipulating your libraries after capture: amplify your post-capture libraries the minimum number of cycles necessary to reach the molarity required by your sequencing facility.


 Bluesky
Bluesky