We generally ask for up to 3-4 weeks after an order is placed to ship myTags libraries.
We recommend using the myTags labeling protocol with myTags immortal libraries for perfroming the labeling process in your own lab. If you would like to use a different protocol, one of our scientists would be happy to provide assistance to ensure success.
Probes are available in 2 configurations to detect both the positive (+) strand and negative (-) strand sequences:
Positive (+) strand probes– SARS-CoV-2 is a class IV RNA virus with an ssRNA genome (positive strand). The probes to detect the (+) strand will hybridize to full-length genomic (+) strand RNA as well as subgenomic (+) strand RNAs made during replication.
Negative (-) strand probes– Detection of the (-) strand is a hallmark of active viral replication. The probes to detect the (-) strand will hybridize to the full-length (-) strand as well as subgenomic (-) strand RNAs. In experimental applications where viral infection is not controlled, additional confirmation of SARS-CoV-2 presence via PCR is recommended.
NOTE: The positive (+) and negative (-) strand probes are not designed to be used in the same hybridization experiment. If co-hybridization is required for your experimental design, please contact us for a customized probe solution.
myTags Expert SARS-CoV-2 probes are available two ready-to-use formats:
- Immortal probes that are ready to be coupled with your preferred tag(s)
- Labeled probes with Alexa-488, ATTO-550, or ATTO-647N fluorescent tags. For experiments that need a brighter signal, the 3X fluorescent tag upgrade is recommended, which can aid detection of lower cellular viral RNA concentrations at earlier experimental time points.
We offer complementary myTags Custom (F)ISH probe bioinformatic design service using our proprietary design algorithms for most types of (F)ISH projects. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will apply our advanced proprietary probe design algorithms to craft a custom probe design for your targeting needs, for any organism or application. Our scientific team has decades of experience designing custom probes for a wide variety of applications, including multi- or single locus/gene localization, chromosome painting, chromosomal indexing/barcoding, haplotyping, and more.
Only the highest specificity regions with minimal background noise and cross-reactivity for premium performance in any downstream experimental application would be selected for the probe design.
And with our transparent and straightforward scientific communication process, you always have full control and ownership over the oligo sequences for each and every myTags Custom probeset.
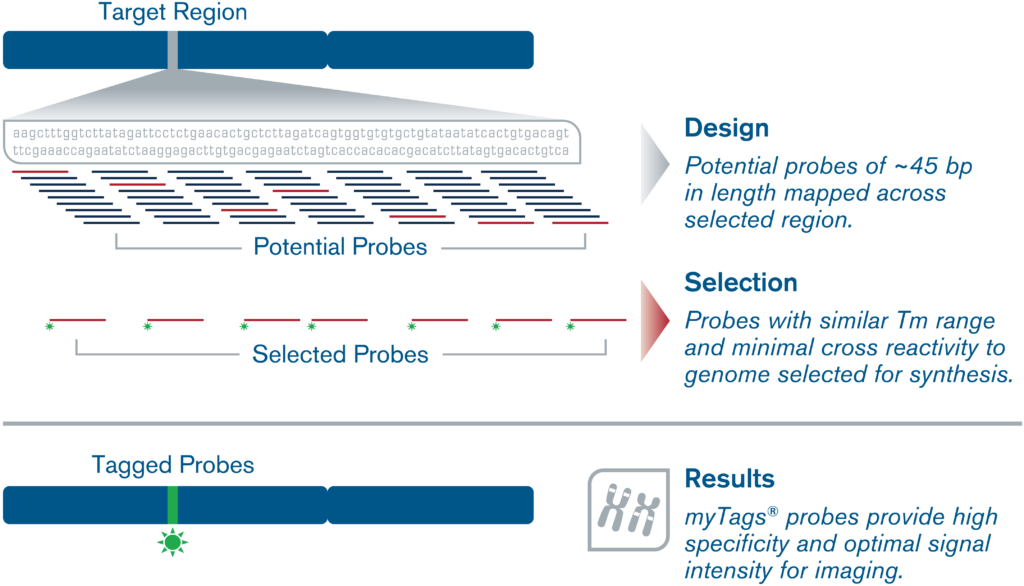
Generally we recommend probe densities between 3-10 probes per kilobase for target regions larger than 50kb. For target regions between 10-50kb, probe densities should be on the higher end of that range, and we may recommend using multiple fluorophores per probe to boost the signal.
We can work with any sequence to design probes. Please contact us with a brief description of your project, including the name of your study species, genomic coordinates, and any additional information.
Our experts will advise you on the most appropriate synthesis configuration for your desired project needs, depending on the number of individual probe sequences required for your design, and the number of individual probesets required for your experiment.
For standalone orders and/or complex probe designs requiring up to 100K+ oligos, our Single Synthesis option provides maximum value. For smaller and/or multiple designs, our new Indexed Synthesis option maximizes both flexibility and cost-effectiveness.
Regardless of the synthesis approach used, all unique probesets are delivered individually and include individual final probeset pool composition verification via next-generation sequencing.
You can choose to receive your individual probe pool(s) either as immortal amplifiable substrate(s) ready for labeling, or pair with our flexible Labeling Service to receive ready-to-use probes for your experiments.
For standalone orders and/or complex probe designs requiring up to 100K+ oligos, our Single Synthesis option provides maximum value. For smaller and/or multiple designs, our new Indexed Synthesis option maximizes both flexibility and cost-effectiveness. All probesets are delivered individually (200 ng minimum yield) and include composition verification via next-generation sequencing to confirm the quality of probe synthesis.
Arbor’s experts will assist you with determining which of our product configurations is most appropriate for your experimental goals and target type(s). You will have full transparency and control over the entire process. Contact us today to get started with your next project and experience the many benefits of using synthetic custom (F)ISH probes.
With Arbor’s flexible oligo synthesis manufacturing technology, we offer two configurations:
Single Synthesis
- Oligo pools containing 1 probeset design
- Can contain up to 108K unique probe sequences per design (larger pools may be available)
- 6 different sizes, depending on total number of probes in design: 1-1.8K, 1.8K-4K, 4K-27K, 27K-54K, 54K-81K, or 81K-108K probe sequences
Indexed Synthesis
- Oligo pools containing 2-10 probeset designs per synthesis event (up to 27K total oligos)
- If >10 designs and/or >27K oligos are needed, add additional oligo pools to scale up
- Oligos are separated into individual probesets via PCR deconvolution
Note: With Indexed Synthesis, short index sequences remain present on the final probe oligos, but are short and not expected to interfere with downstream (F)ISH protocols.
For all myTags Custom orders, final oligo pool composition is verified by next-generation sequencing (NGS), and oligos are delivered as individual probesets.
Add on myTags Labeling Services to get your (F)ISH experiments started quickly!
Ann Arbor, MI 48103
(d/b/a Daicel Arbor Biosciences)
All Rights Reserved.
