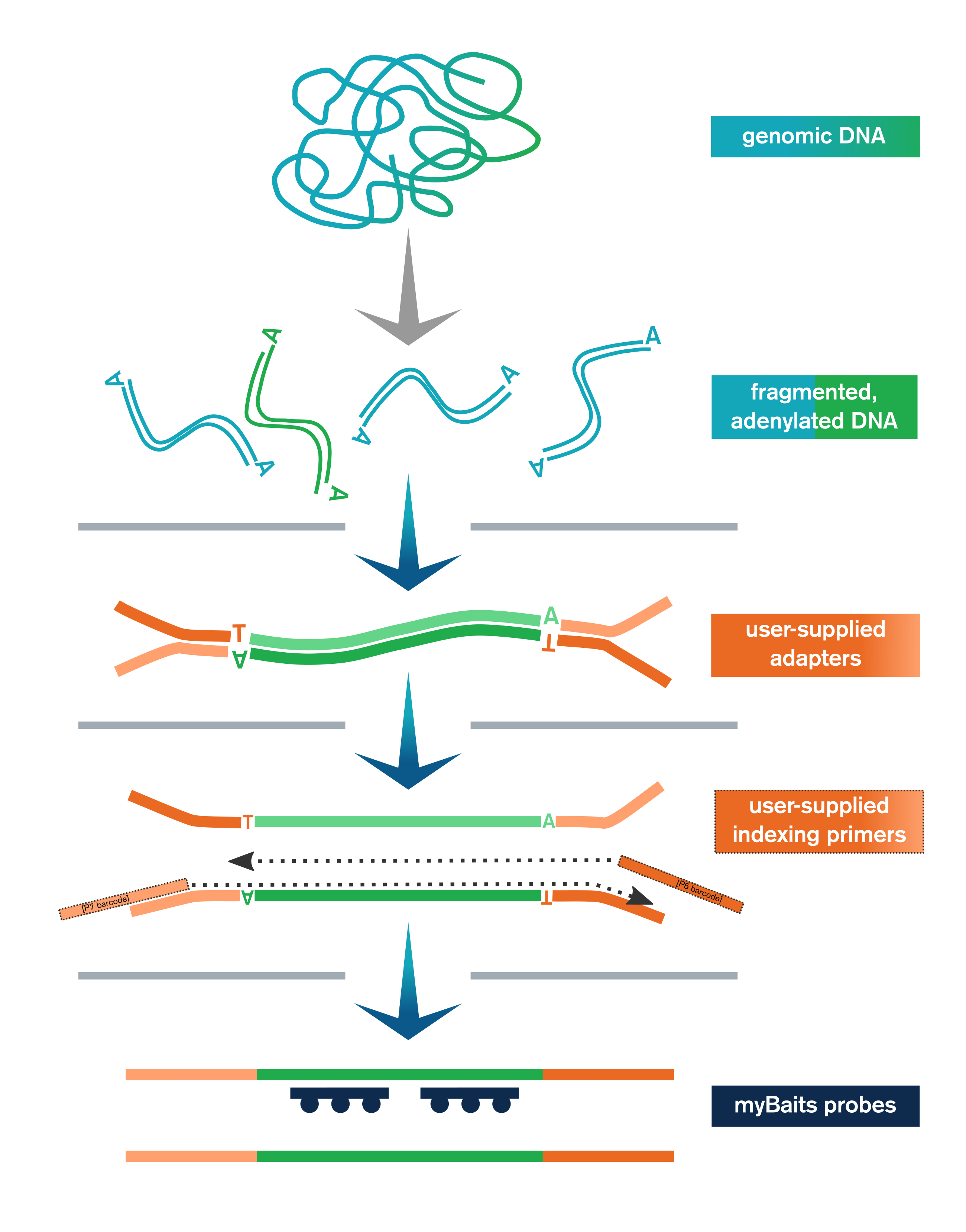
The Library Prep Kit streamlines NGS library preparation for hybridization capture with myBaits. It includes:
- Genomic or other dsDNA is enzymatically fragmented, end-polished, and adenylated
- User-supplied adapters are ligated to the end-repaired fragments
- Ligation products are purified with SPRI beads and then amplified and SPRI’d again
- Libraries are optionally pooled and taken to myBaits capture
In addition to the upstream library preparation steps, the Library Prep Kit for myBaits includes the reagents necessary for performing the post-capture amplification step of the myBaits protocol.


 Bluesky
Bluesky